Time is often our most valuable resource. This post talks about how to shave time off your amino acid analysis.
I’ve got a third installment for our mini-series on the AdvanceBio Amino Acid Analysis discussion. We’ve already covered preventative tips to keep things running smoothly and troubleshooting common problems with this method, now let’s talk about how to save ourselves a few minutes of analysis time. For one or two samples, shaving a couple minutes off the run time may not make a big difference, but it’ll really add up in time and mobile phase savings the more samples you have to analyze.
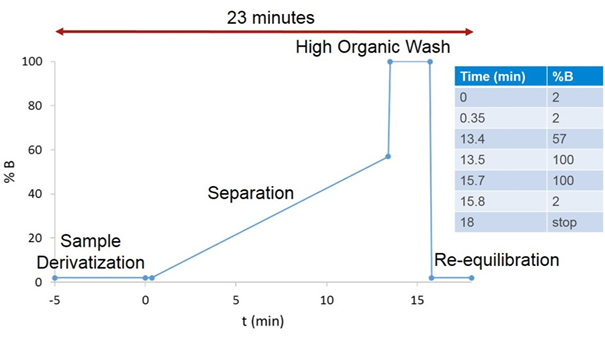
Here’s the gradient profile for the amino acid separation, with time for the amino acid derivatization added at the beginning (this 5 minutes is an estimate – each autosampler model takes a slightly different amount of time):
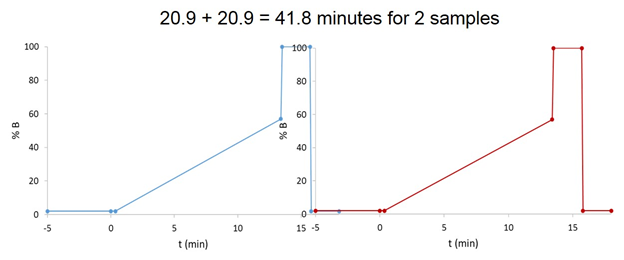
As illustrated (and written in the “How-to” Guide), it would take ~ 46 minutes to run two samples. However, during the sample derivatization, the autosampler is in bypass mode, so the initial mobile phase conditions are flowing through the column – that is, the column is undergoing additional equilibration while the sample is equilibrating. Instead of ending the method at 18 minutes, we end it at 15.9 minutes, immediately after returning to the initial mobile phase conditions, we save just over 2 minutes per injection. Sure, I’m generally not worried about another 2 minutes for a single sample, but to run calibration standards and a large sample set, that time and mobile phase adds up.
That’s it for amino acid analysis for now. Talk soon!
- Anne
Keywords: Bio columns, liquid chromatography, tips and tricks, amino acids, AdvanceBio blog

-

cptan
-
Cancel
-
Vote Up
0
Vote Down
-
-
Sign in to reply
-
More
-
Cancel
Comment-

cptan
-
Cancel
-
Vote Up
0
Vote Down
-
-
Sign in to reply
-
More
-
Cancel
Children