An introduction to a salty complement to reverse phase chromatography to add to your analytical toolbox
Hello everybody! This week kicks off a series of blog posts on a topic that’s completely new to me. I’ve therefore asked some of my colleagues to contribute while I learn alongside you. My colleague Greg from our R&D team is going to get us started.
---
This blog series will tell you everything you need to know (I hope!) about a mode of chromatography that you might not be familiar with: hydrophobic interaction chromatography (HIC). If you’ve never used HIC in your lab, you’re not alone. HIC is not nearly as common as other separation methods such as size exclusion or reversed-phase (RP). However, HIC is becoming a lot more popular, as evidenced by the increasing number of conference and literature presentations on the subject. The reason? Increasingly complex protein therapeutics continue to put our analytical techniques to the test, and HIC is a valuable option for taking on these separation challenges.
I’ve always enjoyed the history of science and I like to kick off learning about unfamiliar techniques by investigating their origins. The term “hydrophobic interaction chromatography” didn’t appear until 1973 [1], but the groundwork for this separation mode was first reported in 1948 [2]. At the time, it was well known that proteins can be precipitated using high concentrations of salts. The new twist was the observation that proteins adsorbed strongly to sorbents for which they would ordinarily have only weak affinity at salt concentrations much lower than would normally be required for precipitation. The first appearance of HIC in a chromatographic format was in 1962, when serum proteins were separated on cross-linked dextran [3]. Materials substituted with hydrophobic functionality appeared in the early 1970s [4, 5], with sorbents offering, “pure” hydrophobic interaction (without any ion exchange properties) appearing in 1973 [6].
HIC has advanced tremendously since it was first described and is often used for large-scale protein purification. In this case, HIC is useful for removing protein aggregates and contaminants such as host cell proteins [7, 8]. HIC is also used for small scale analytical separations, which will be the focus of our blog series. Due to its unique selectivity, HIC is complementary to techniques like size exclusion, ion exchange, and RP. Of these techniques, HIC is most like RP, in that separation is strongly correlated with protein hydrophobicity. But there are two key differences between HIC and RP: First, HIC separations are driven by a decreasing gradient of salt, rather than an increasing gradient of organic solvent as in RP. Second, HIC separations maintain the native state of a protein, and thus the protein remains biologically active following the separation. This activity allows collection of purified peaks for downstream potency assays, for example. Furthermore, the native state of the protein means that features on the surface of a protein tend to drive selectivity in HIC mode. Therefore, HIC is particularly suited to separating post-translational modification or degradation products of a protein, which is sometimes difficult to achieve by other modes of chromatography.
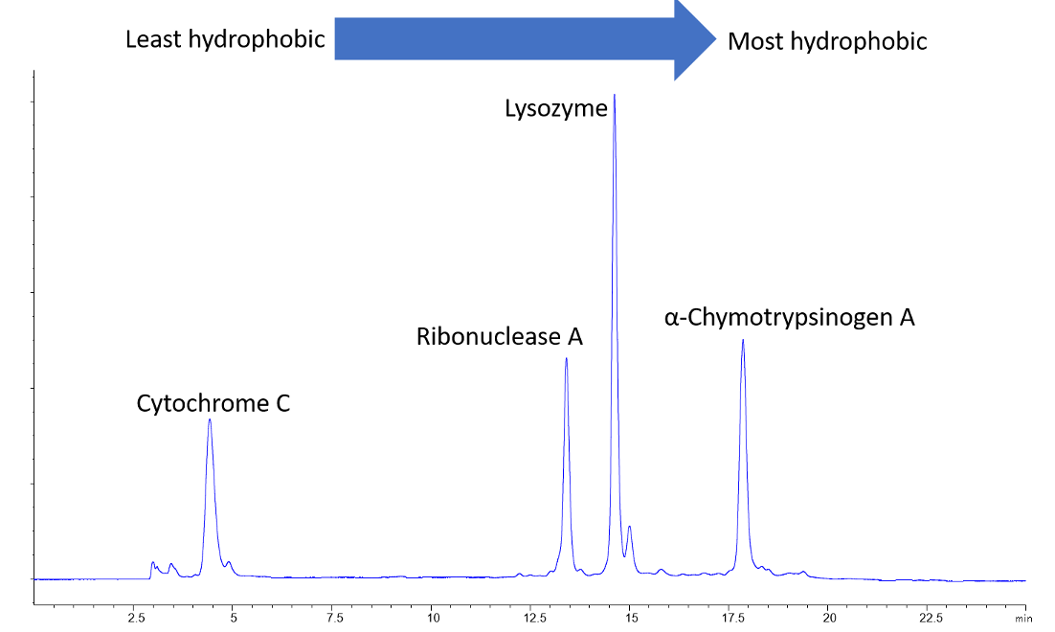
We should spend a little time addressing the topic of salts. Initial conditions for HIC separations start with a very high concentration of salts called lyotropic salts. These salts, which are unique in their ability to “salt out” proteins, are conveniently summarized in the Hofmeister series. The most commonly used lyotropic salt in HIC is ammonium sulfate, and separations typically start with concentrations in the 1–2 M range. The concentration of ammonium sulfate is gradually reduced over the course of the gradient, and proteins are separated according to their relative hydrophobicity (the more hydrophobic, the later the RT - thus lower concentrations of salt are required for more hydrophobic proteins). There’s been considerable work on understanding the mechanism of HIC separations. If you’d like to read a summary on some of the investigations into protein behavior on HIC, I’ll point you to a review by Lienqueo [9].
Proteins elute in order of increasing hydrophobicity as the salt content of the mobile phase decreases.
So how about some recent examples of how HIC is being used for biologics characterization? One of the main success stories relates to antibody drug conjugate (ADC) characterization [10]. In this case, HIC can be used to separate ADCs based on the number of conjugated small molecule drugs, and is thus useful for calculating the drug to antibody ratio or DAR [11]. HIC has also been shown to separate Asp/IsoAsp isomers as well as succinimide variants [12] and oxidation variants of methionine and tryptophan [13]. Bispecific mAbs, a modality of increasing importance, can be analyzed using HIC, in particular for assessing the population of parental mAbs versus the intended bispecific product [14]. Relative hydrophobicity of proteins can also be assessed, as a potential indicator of aggregation [10].
Methionine oxidation and aspartic acid isomerization are two of the post-translational modifications monitored by HIC.
I hope this introduction gives you a taste of what HIC has to offer and that you enjoy this blog series. I think you’ll find there’s lots of reasons to add HIC to your analytical toolbox if you haven’t already. The advent of so many new therapeutic modalities will surely provide some opportunities to shine for this powerful mode of separation.
--
Hi all, Anne here again. Thanks, Greg! Over the next few weeks we’ll expand upon some of the topics Greg touched on – salts and mobile phase preparation, method development for ADCs and other proteins, and some best practices to stay out of trouble while using high salt mobile phases. Until next time.
Anne
References:
- Hjerten, S., Some general Aspects of Hydrophobic Interaction Chromatography. J Chromatogr, 1973. 87: p. 325-331.
- Tiselius, A., Adsorption Separation by Salting Out. Arkiv foer Kemi, Mineralogi och Geologi, 1948. 26B: p. 1-5.
- Porath, J., Zone Precipitation. Nature, 1962. 196: p. 47-48.
- Yon, R.J., Chromatography of Lipophilic Proteins on Adsorbents Containing Mixed Hydrophobic and Ionic Groups. Biochem J, 1972. 126: p. 765-767.
- Er-el, Z., Y. Zaidenzaig, and S. Shaltiel, Hydrocarbon-coated Sepharoses. Use in the Purification of Glycogen Phosphorylase. Biochem Biophys Res Commun, 1972. 49: p. 383-390.
- Porath, J., et al., Salting-out in Amphiphilic Gels as a New Approach to Hydrophobic Adsorption. Nature, 1973. 245: p. 465-466.
- Jiang, C., et al., Defining process design space for a hydrophobic interaction chromatography (HIC) purification step: application of quality by design (QbD) principles. Biotechnol Bioeng, 2010. 107(6): p. 985-97.
- McCue, J.T., et al., Modeling of protein monomer/aggregate purification and separation using hydrophobic interaction chromatography. Bioprocess Biosyst Eng, 2008. 31(3): p. 261-75.
- Lienqueo, M.E. and A. Mahn, Predicting Protein Retention Time in Hydrophobic Interaction Chromatography. Chemical Engineering & Technology, 2005. 28(11): p. 1326-1334.
- Fekete, S., et al., Hydrophobic interaction chromatography for the characterization of monoclonal antibodies and related products. J Pharm Biomed Anal, 2016. 130: p. 3-18.
- Wakankar, A., et al., Analytical methods for physicochemical characterization of antibody drug conjugates. mAbs, 2014. 3(2): p. 161-172.
- Valliere-Douglass, J., A. Wallace, and A. Balland, Separation of populations of antibody variants by fine tuning of hydrophobic-interaction chromatography operating conditions. J Chromatogr A, 2008. 1214(1-2): p. 81-9.
- Boyd, D., T. Kaschak, and B. Yan, HIC resolution of an IgG1 with an oxidized Trp in a complementarity determining region. J Chromatogr B Analyt Technol Biomed Life Sci, 2011. 879(13-14): p. 955-60.
- Zwolak, A., et al., Rapid Purification of Human Bispecific Antibodies via Selective Modulation of Protein A Binding. Sci Rep, 2017. 7(1): p. 15521.
Keywords: Bio columns, liquid chromatography, hydrophobic interaction chromatography, HIC, AdvanceBio blog