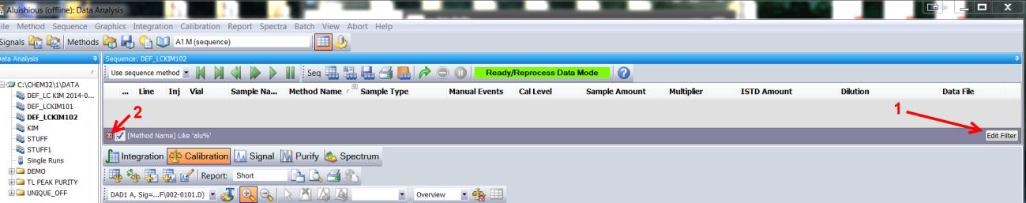
In OpenLAB CDS Chemstation, you can filter the Navigation table using the Filter Editor. For example, you may only want to look at Calibration Stds, so you filter on Sample type. However, if the Data set you are loading does not comply with the filter, you will have an empty table when you load the data.
Each column could possibly have a filter. If a filter is applied to a column, you will see a little icon in the upper right of the column header that looks like a capitol Y with a line across the top.
Here are two indicators that a filter has been applied:
- You load your Results Set/Sequence Container by double clicking on it in the Navigation Tree, but not all of the data files load into the Table.
- "No signals available" message appears when loading a Results set
To resolve this issue, remove the filter from the column. To remove the filter from your table, you must delete the filter. You can do this two ways. One, click on the filter editor button at the bottom right of the navigation table, then delete the entry. Or Two, click on the red x at the lower left of the Navigation Table at the beginning of the Filter line.
Once the filter is removed, data will load correctly into the Navigation Table.
42130
2015-0512, 2016-0527