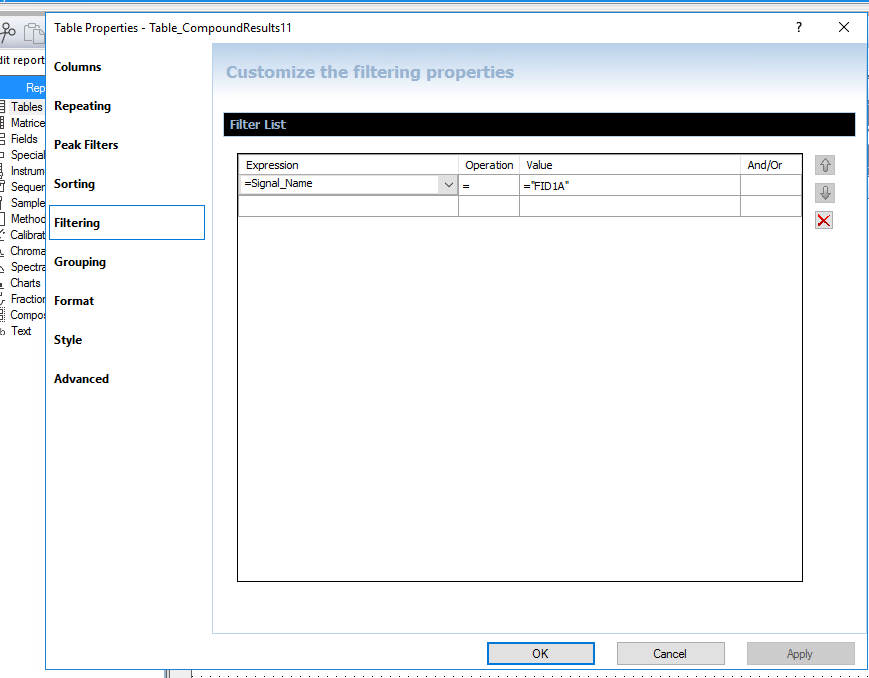
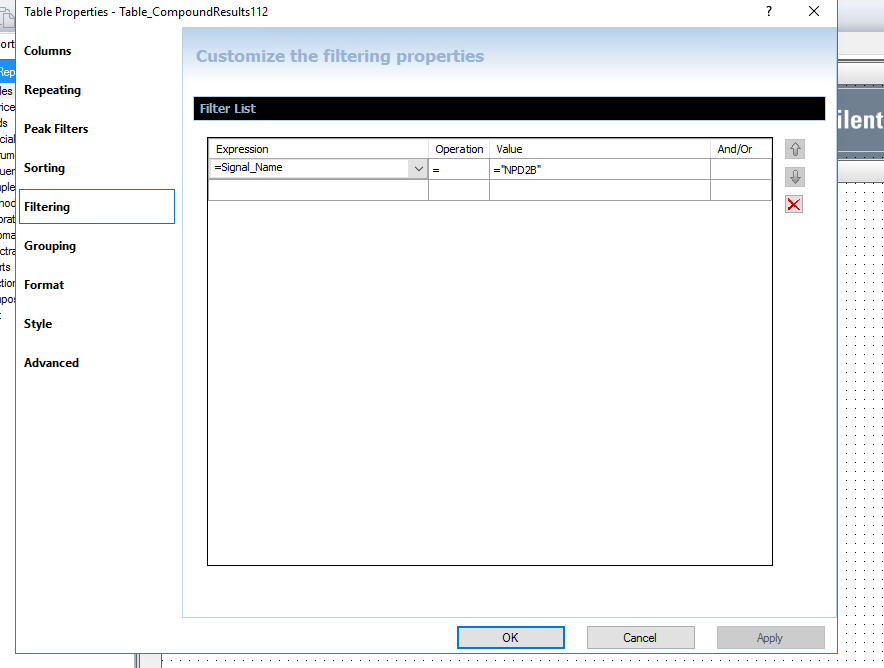
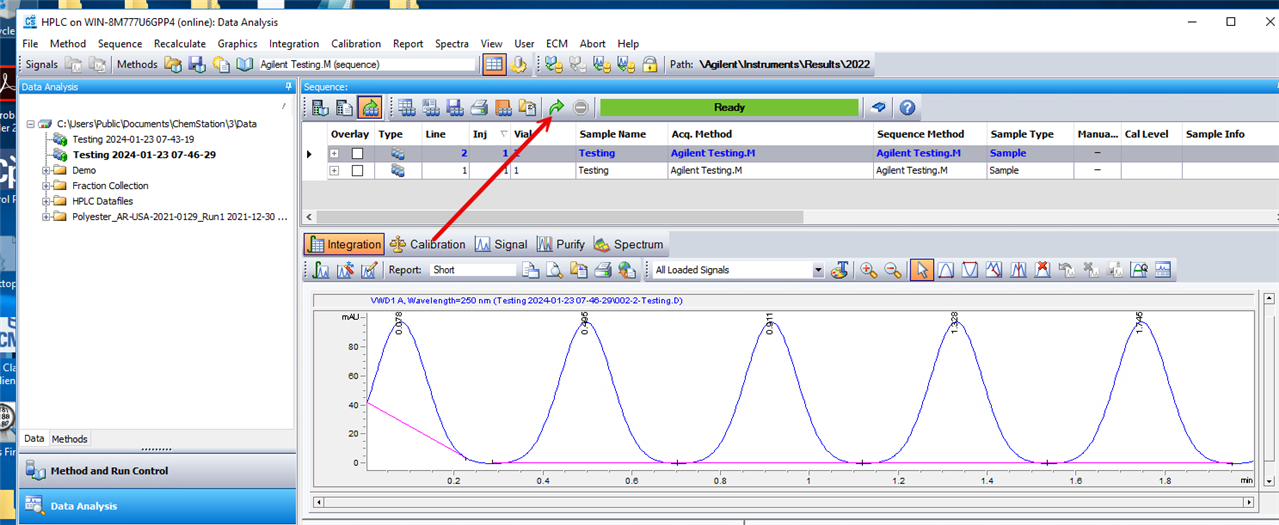
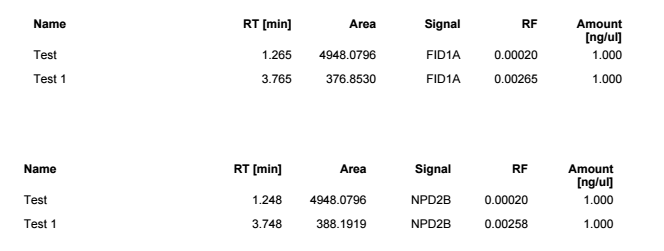
I am using OpenLab Chemstation C.01.10 on an 8890 with dual FIDs. I was trained on HP's version of Chemstation that reported both detector quant amounts in one row for precision purposes. It made it 10x easier to review data and confirm sample concentrations for my HAA5 analysis with both signals on the same row. I have been fiddling with the report designer, but I cannot seem to figure it out. Is it possible for the software to report in the manner described?


 \
\