Hello everyone,
It is my first time using MassHunter Quant (version 12.0), and I have some questions regarding setting up parameters in quant methods. These questions might be very basic. Thank you in advance for the help.
Question 1:
Is there a way to integrate all peaks within one acquisition window, and calculate the concentration of each peak without having to define each individual compound please?
In the attached screenshot, there are over ten target peaks in one window. The predominant peak in green is a toxic compound, included in the calibration curve using the average RF method. The other peaks, non-toxic compounds, do not require individual naming, but their concentrations are calculated using the RF of the toxic compound in the same homologue group. Rather than setting up over 10 single compounds individually, assigning them with the same compound group and adding them into a total compound, is there a way to establish a total compound only and have the software integrate all peaks within a specified RT range?
Question 2:
How can integration parameters be configured to include multiple coeluting peaks as one peak?
Some analytes are coelutions, so we need to include multiple peaks as one peak as shown below. Is there a way to set up integration parameters to achieve this and save time on manual integration please?
Question 3:
Is it mandatory for a Qualifier to exist in the method?
We have some targets that are known with very low responses for the confirmation ion, and therefore, we ignored them in our SOP. Is there a way to delete the Qualifier row in the quant method?
Question 4:
How does the software calculate noise if “Noise Regions” column is left blank? Will the software automatically identify a representative piece of noise within the acquisition window?
Question 5:
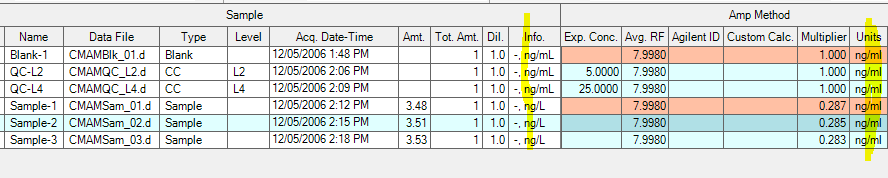
How to calculate sample-specific detection limit (SDL) for each analyte please?
Our SDL formula is SDL = a factor (2.5 or 3) * Noise * average RF of that compound from the Calibration Curve.
Which column should be used for this calculation?
Any help would be much appreciated!