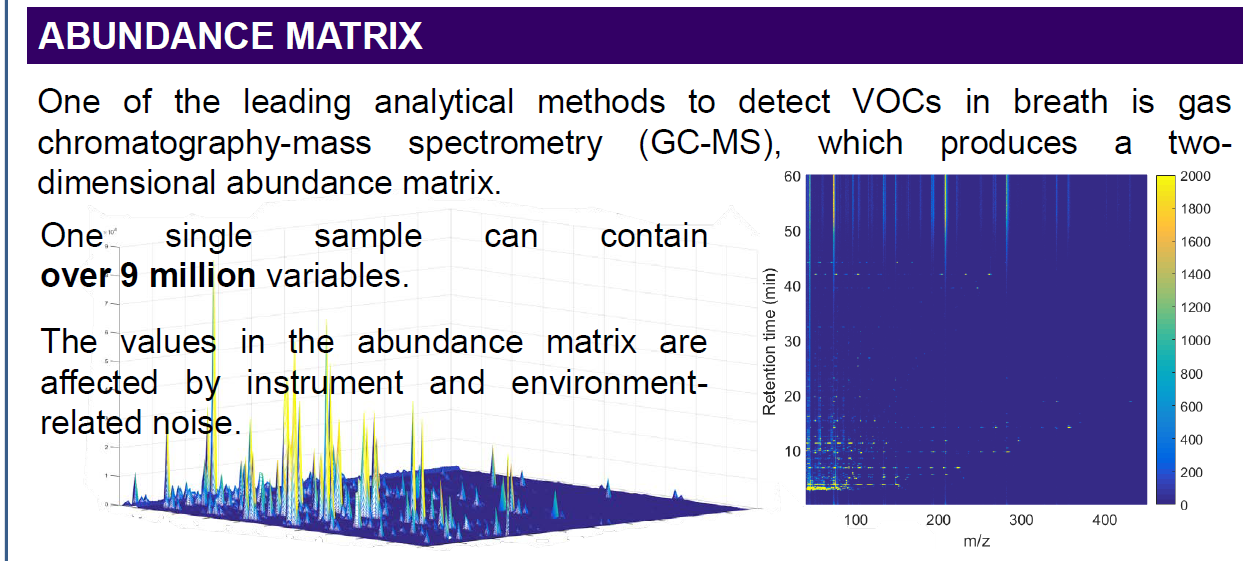
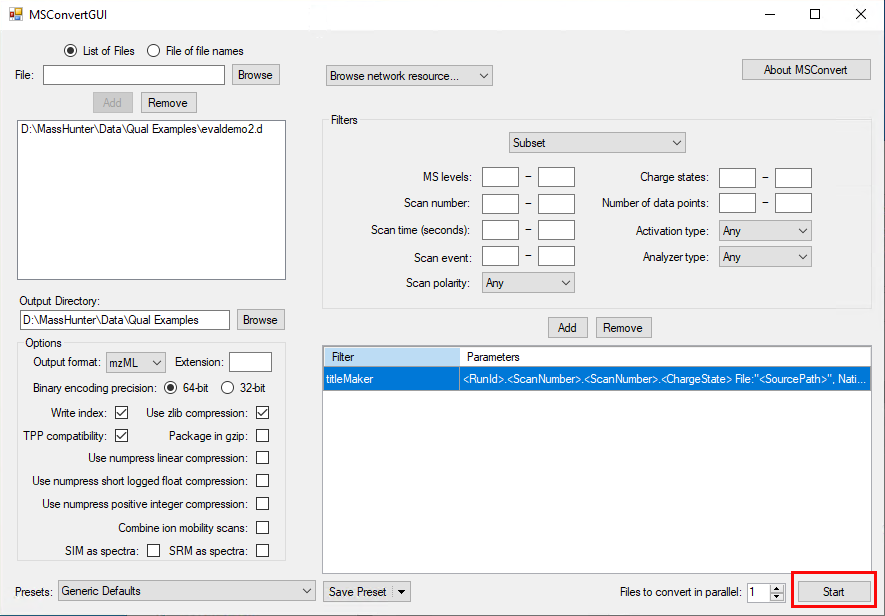
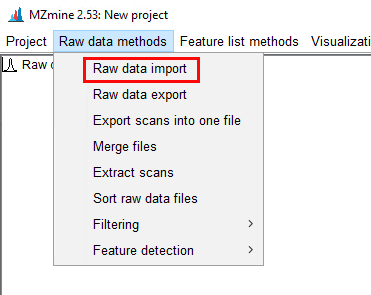
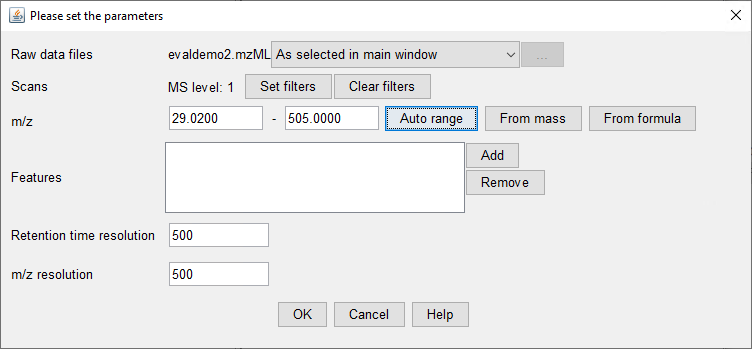
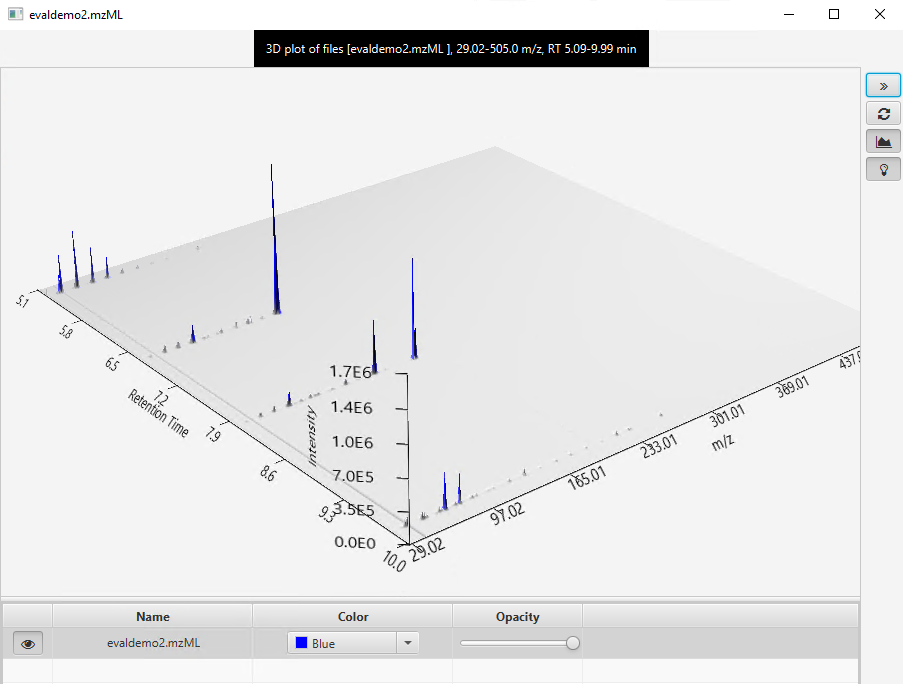
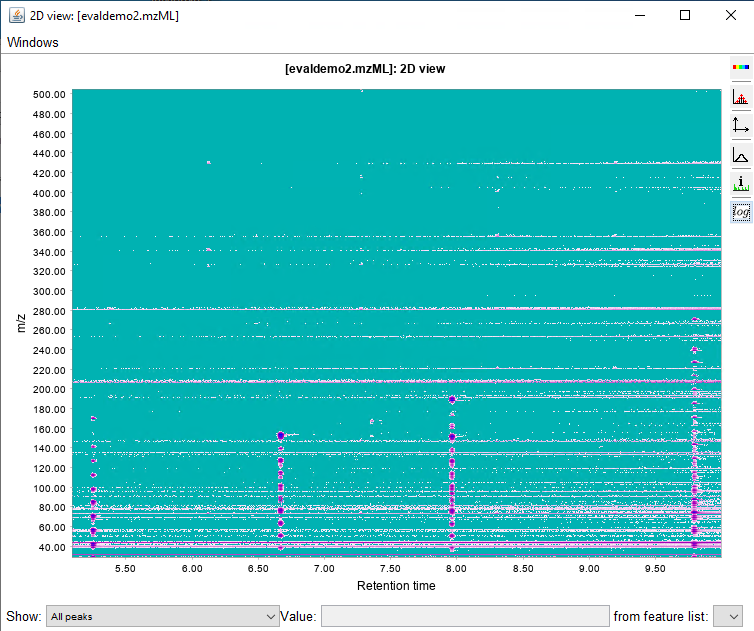
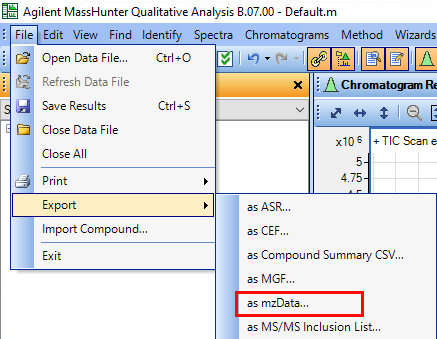
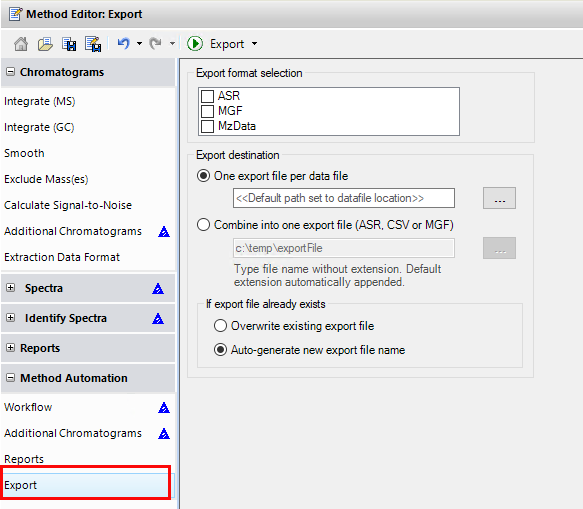
I would like to get the raw data from an unkown sample chromatogram which is in SCAN mode, so I can be able to build this 3-Dimensional plots, for example: In X axis there are all of the m/z values; in the Y axis the Retention Time and the results would show the intensity/abundance for every m/z in their respective RT.
I want to know if this is possible using Data Analysis Software or Mass Hunter. Besides it would be helpful to get a step by step guide.
I really would aprecciate it!