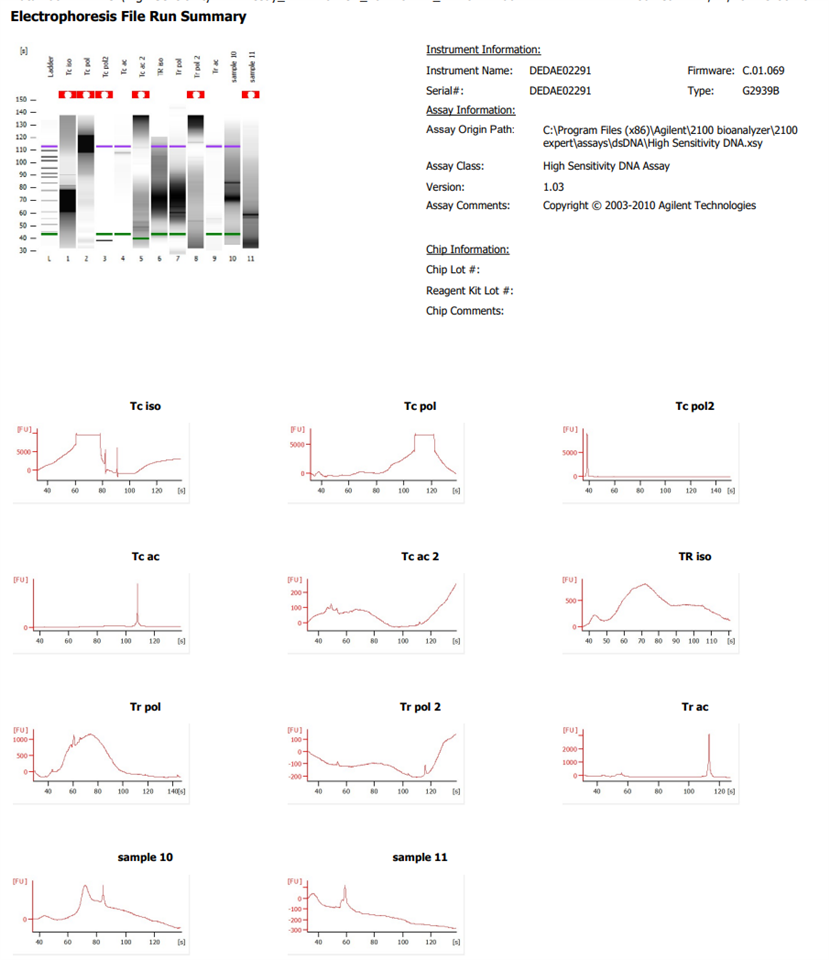
Hello,
I have analysed some samples from CUT&TAG previously with good results. However, now the results I obtained from the bioanalyser 2100 show a really strong signal (the optic signal too high error) as well as weird gels with even no markers. I have tried DNA extraction with phenol or with beads to see if any contaminant was the problem but the analysis obtained from the instrument is still not better. The samples are diluted in nuclease-free water. I have already redone the gel and changed the syringe in the priming station.
What else could be affecting the samples to obtain these strange results when it used to work previously, following the same protocol?