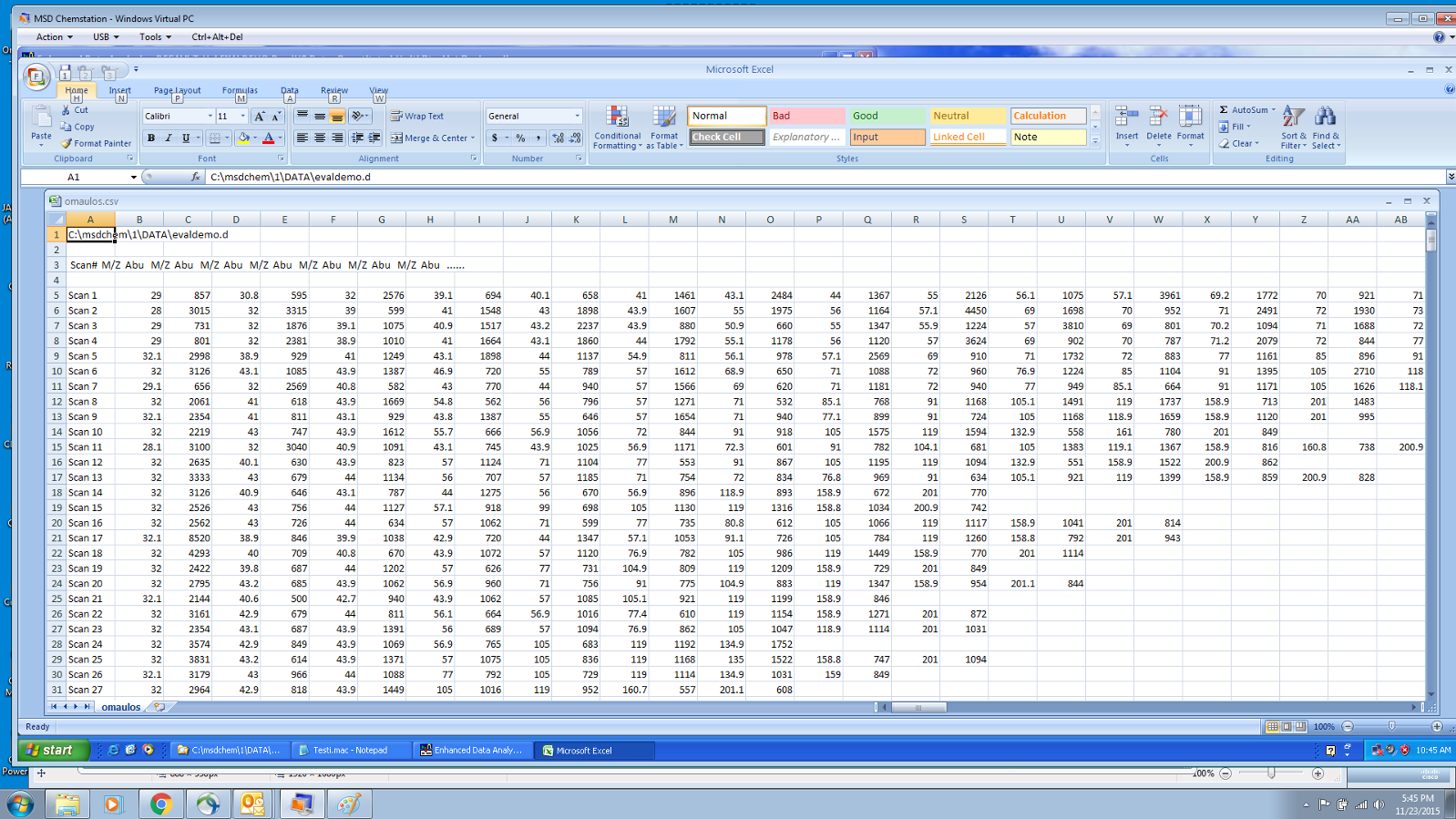
Hi All
I have the output of several thousand organic acid MS procedures and need to import the data into a structured database for further analysis. The files I have available are xls (as attached) and data.ms. As far as I know these are the output from an AGILENT 5975C & 7890A mass spec.
I was expecting to find data that gives me abundance vs m/z ratio i.e. simple xy values but the xls files have several data points for each compound.
- Is there documentation available that describes the xls format?
- Is there a software library that allows me to import the data.ms files so I don't have to parse the xls files?
BTW there are also a whole bunch of other files such as ini, res and txt files.
Any advice appreciated.
Thanks.