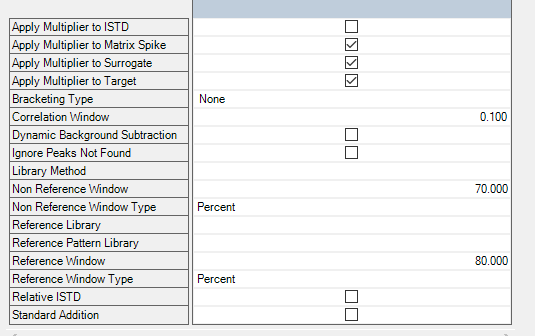
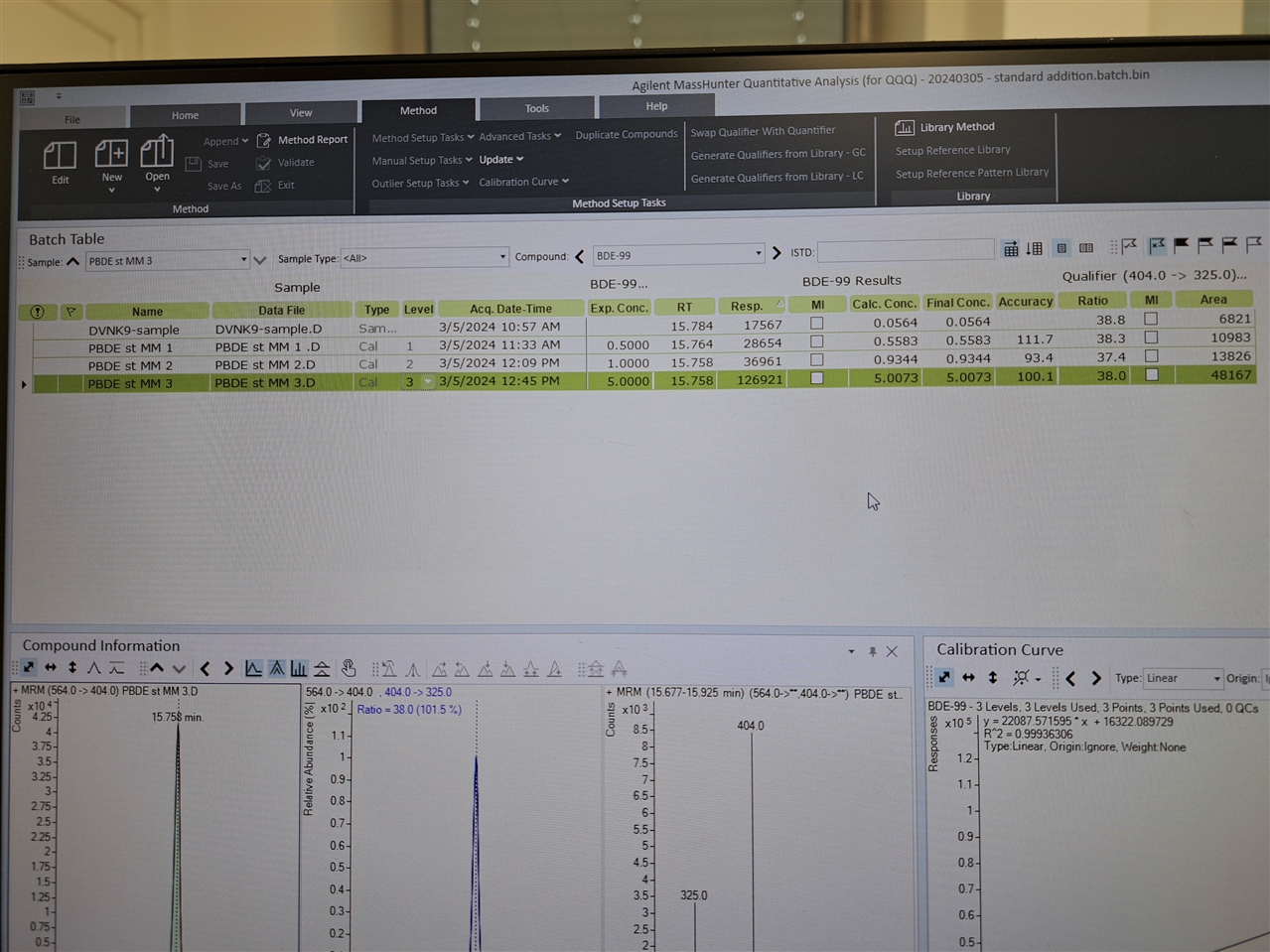
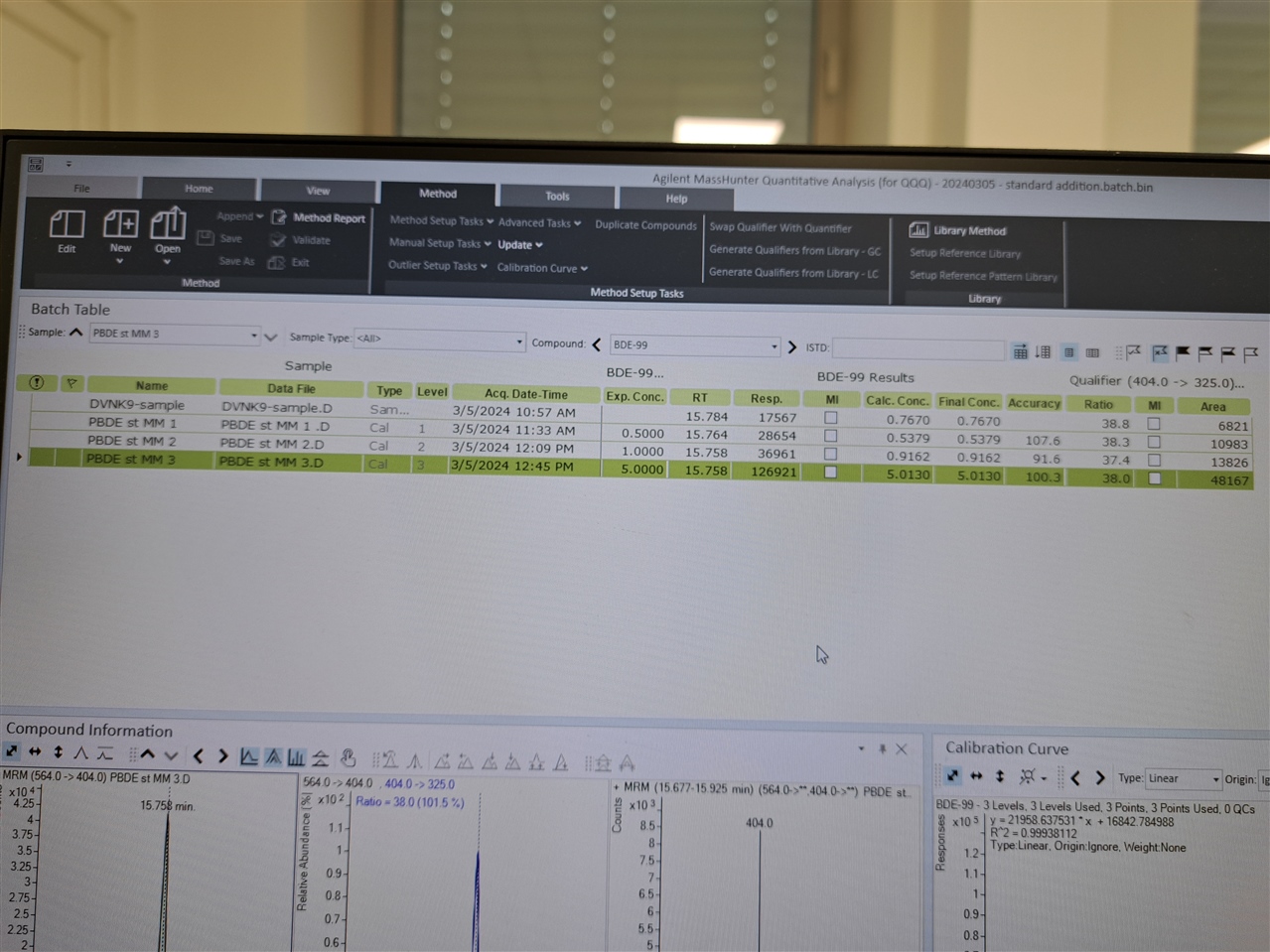
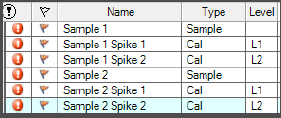
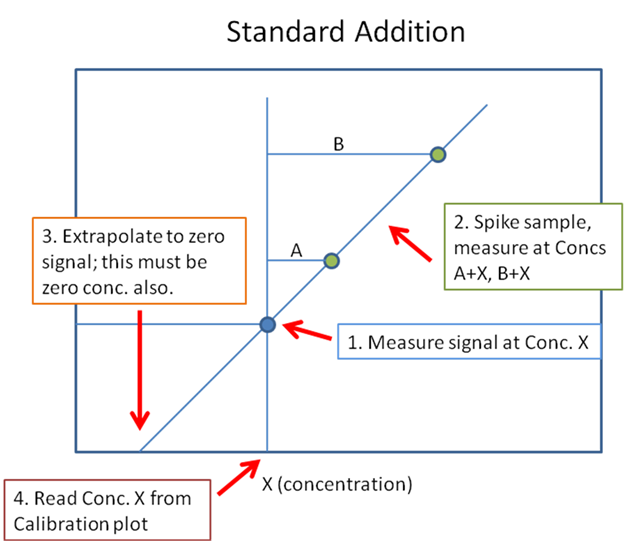
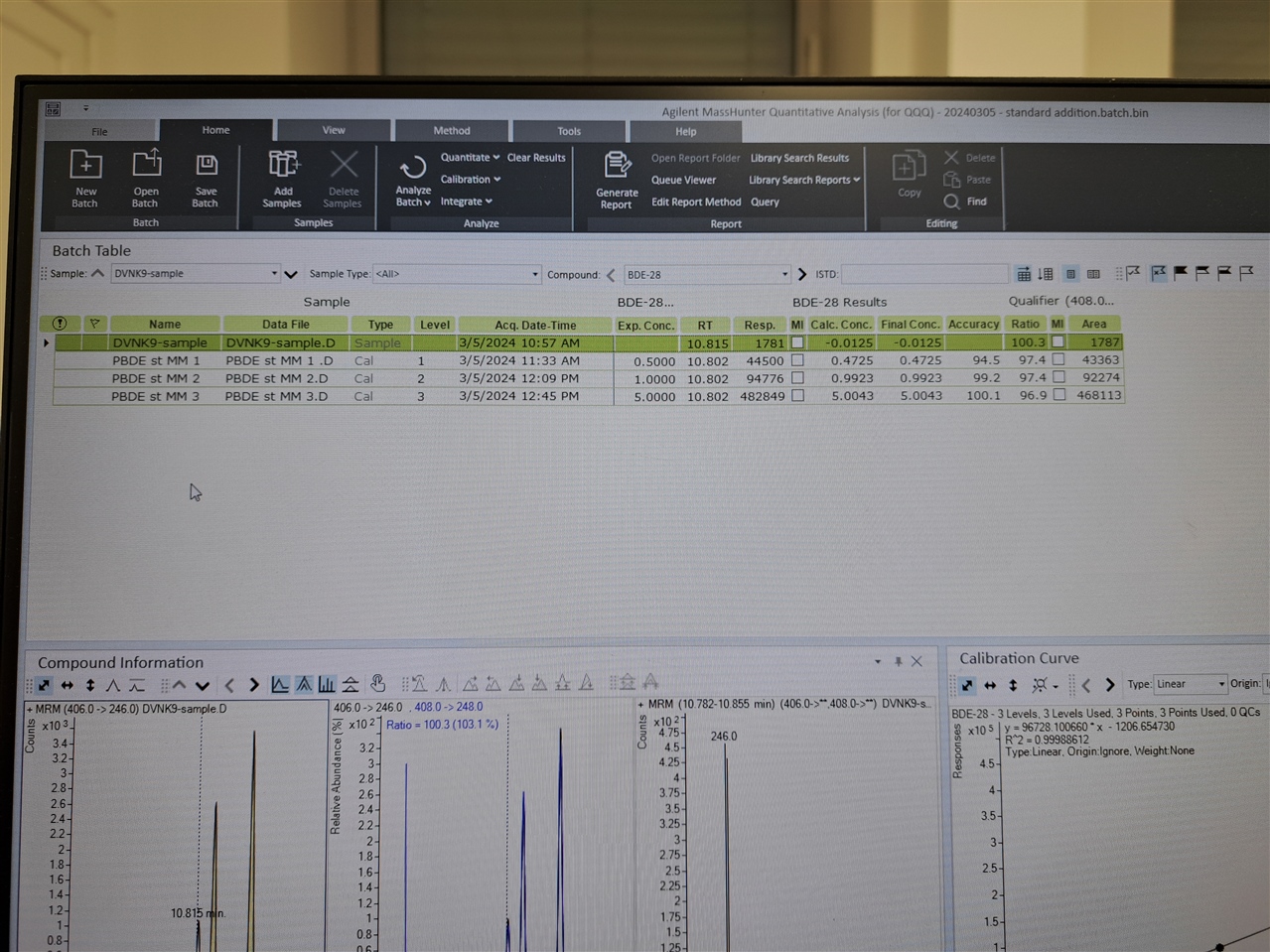
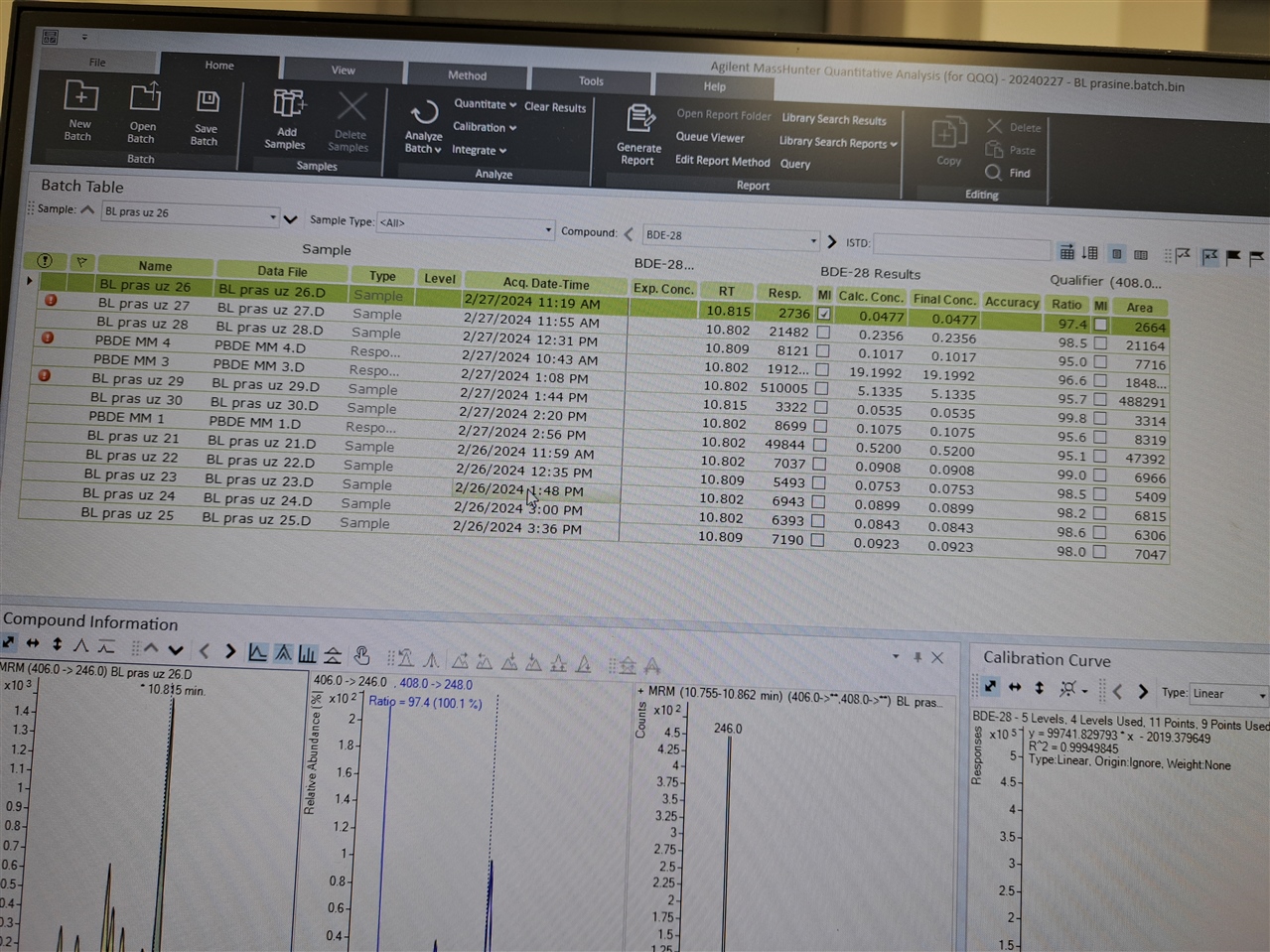
Hello, everyone! I need software setup instrucitons (Mass hunter 10.2; GC-(QQQ)MS/MS) for using matrix matched standards in which we have some of our analytes present in matrix. So, we have matrix (where we detected 2 out of seven analytes), and we used that matrix for setting up our calibration curve.