Hi everyone,
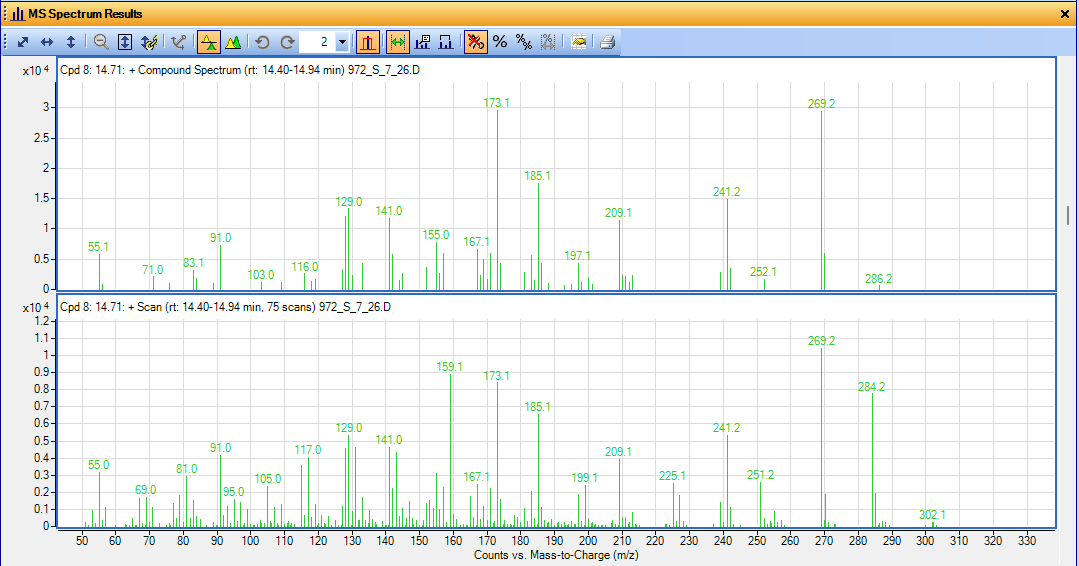
I have problem with MassHunter Qual in finding compounds in my sample. It's really frustrating. When I click on "Run Compound Automation Steps" the resulting compound spectra are way different than the raw scan. And because of that, it can't find any hit in the library. However, when I run manual spectral search I can find good hits. They problem is that MassHunter removes my key spectra for no reason, and I can't figure out how to correct this so that I can get what I need in the "Compound list" and export all the dataset in one file (I have a total of 480 samples). Here is a screenshot of the "Compound Spectrum" and "Scan" for the same peak. Compound spectrum doesn't lead to any library hit, but I can manually find great hits. My samples are pine resins and peak 284 is the molar mass of my intact molecule of diterpene resin acid. Is there a way I can prevent MassHunter from dropping my spectra and at the same time, give me a compound list? I can just rely on scan results but they don't show up in the compound list so I have to export each 480 sample manually to csv file and then merge all of them which is a pain.
Any help is greatly appreciated!
Thank you!