Hello,
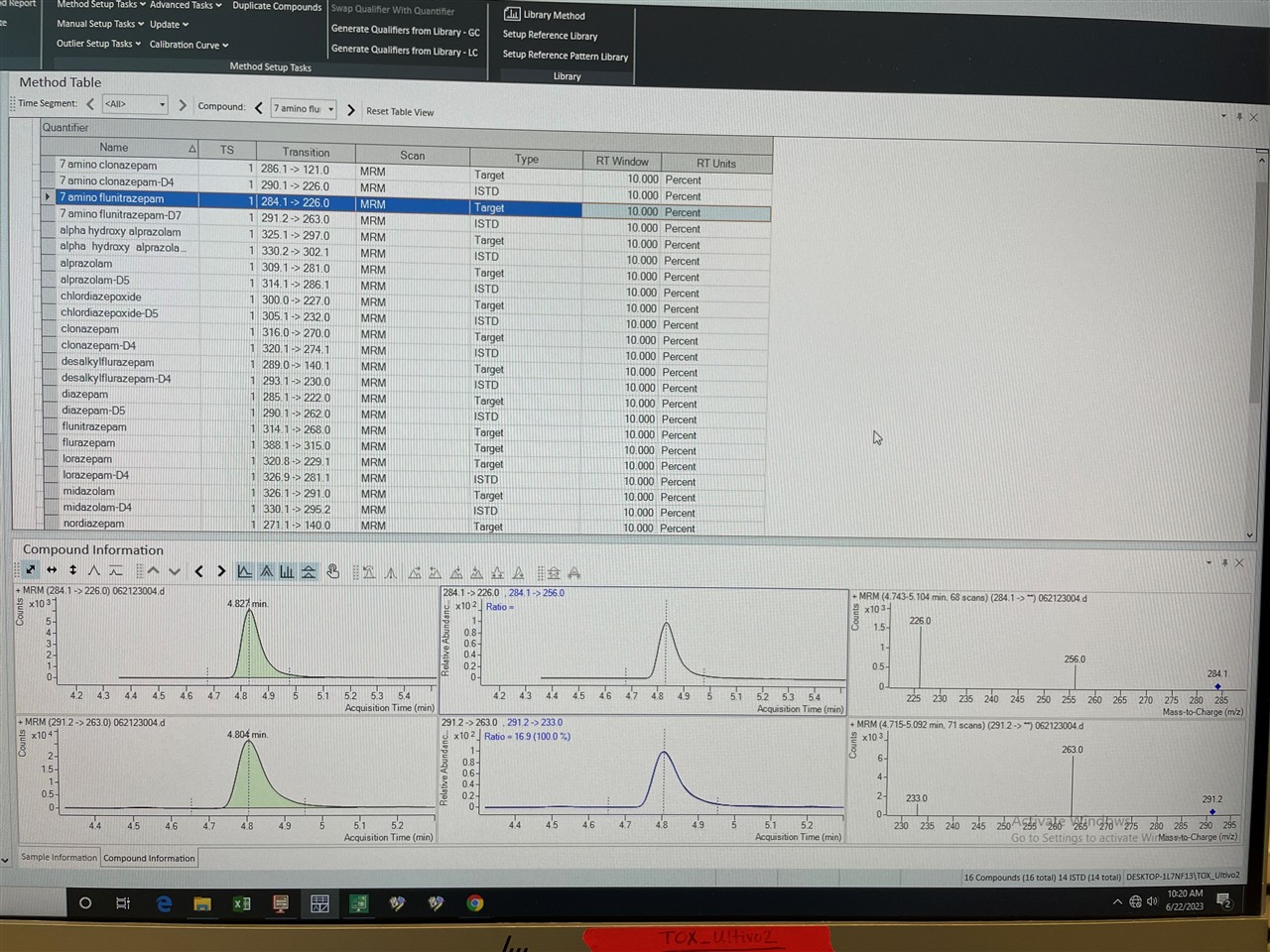
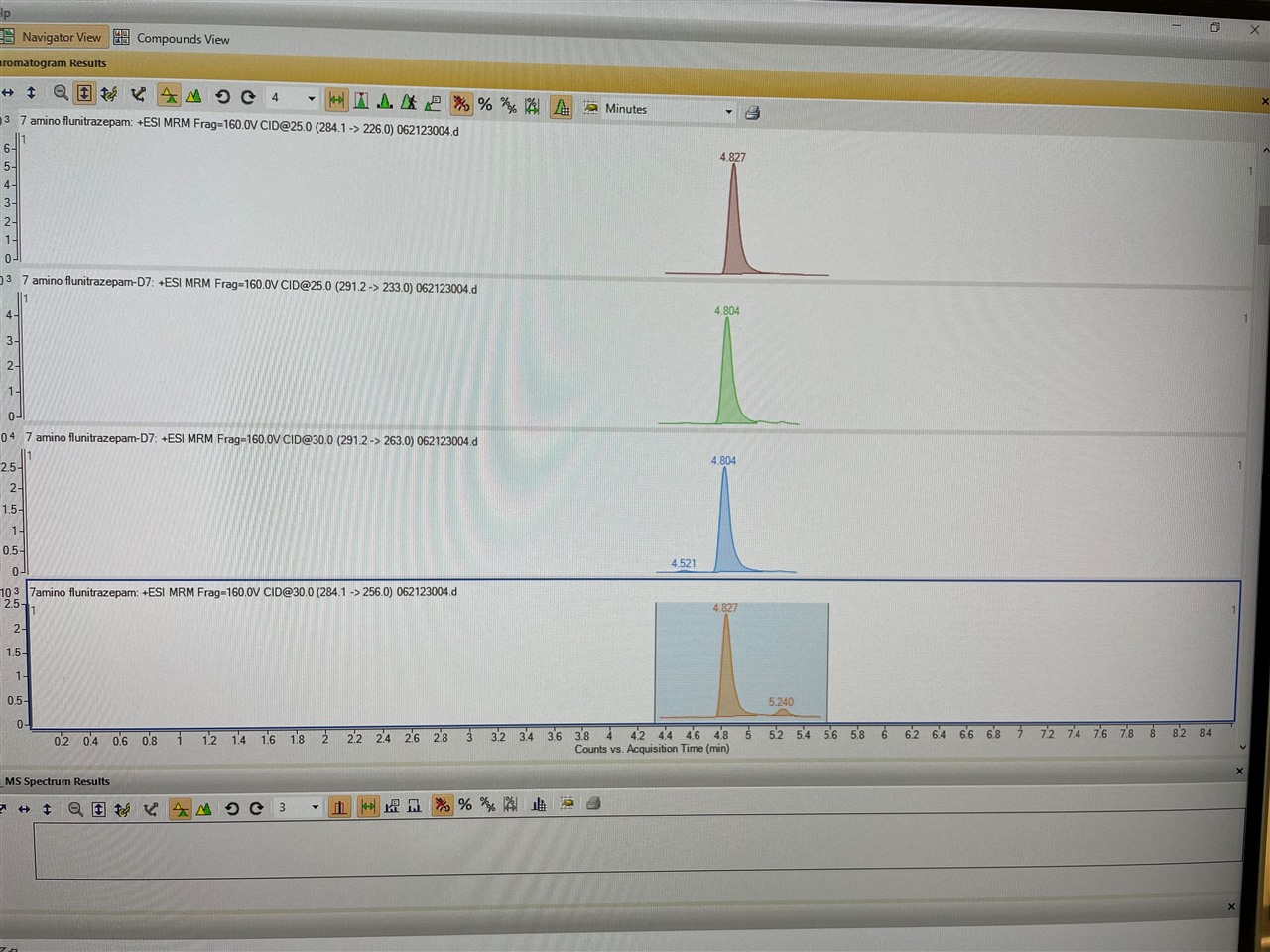
We have been having issues with MassHunter Quant Software (version 12.0) randomly not finding qualifier peaks (284.1 -> 256.0). The TIC shows that the ion is there and was extracted (see screenshot, right-most panel), and when we open up the data files in the Qualitative software the peak is found at the indicated RT (see screenshot, highlighted peak). We have tried re-building the method from scratch to no avail. Of note, when we analyze the data in an older version of MassHunter Quant it integrates the peak just fine. All of the integration and quant method parameters are identical.