Hi
We've had this issue for a while. There is one compound in this analysis. While its RT is at 6.24, there will always be a signal drop at 6.3. The signal drop looks like a negative peak from the base line.
Here is what we noticed:
In blanks samples: no drop;
Junk peak with same transitions but different RT: no drop;
Different transition but with same parent ion, same RT: same drop;
Different transition but with different parent ion, same RT: no drop;
Other analytes in the same method, different parent ion different RT: no drop.
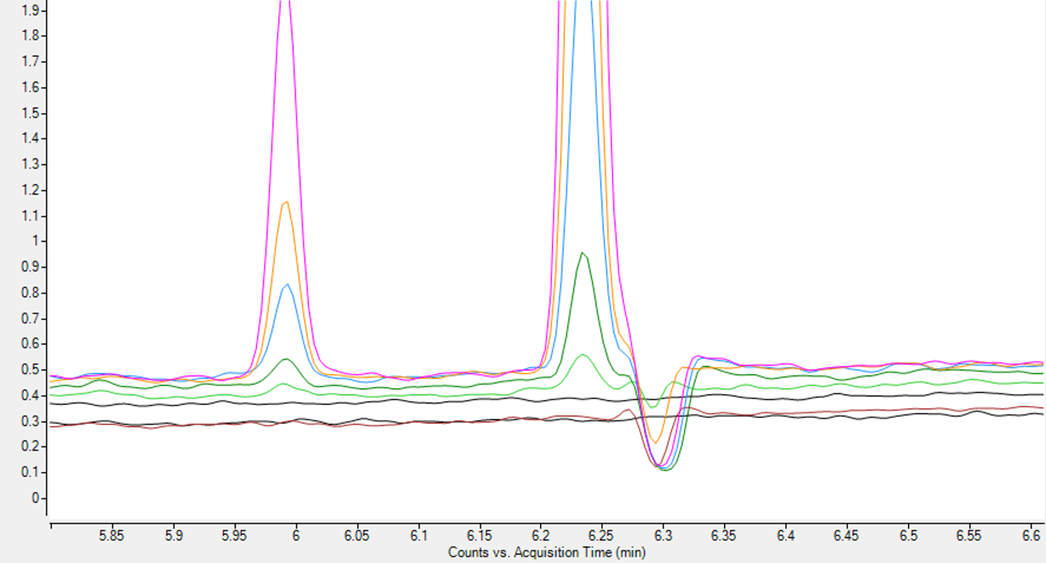
This figure shows the drop in a bunch of std curves. Two black lines are blanks.
I have also ran a MS2 scan and saw no interference in that 6.3min area. What do you think could be the reason? The flow is 1 mL/min, pressure is 5psi in the beginning condition, column at constant flow. Tune seems ok to me as well (pass check tune and EMV not high <1100, 69 at good abundance10,000,000-ish). Ion source has just been cleaned.
Thank you!