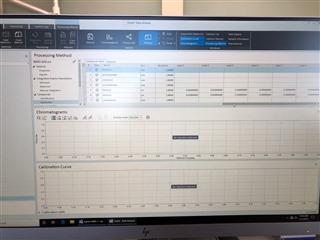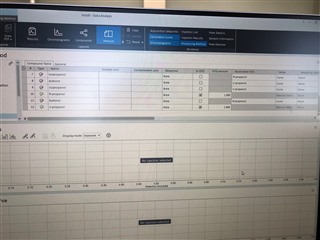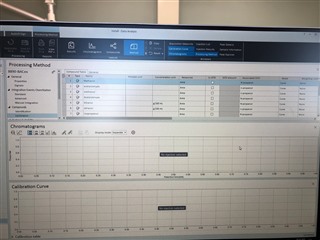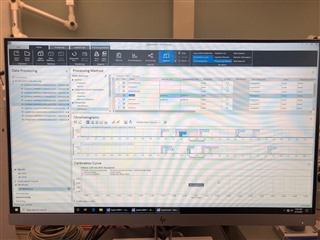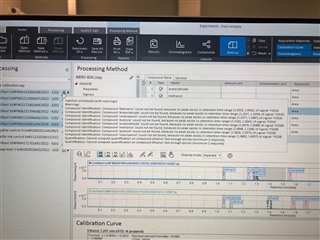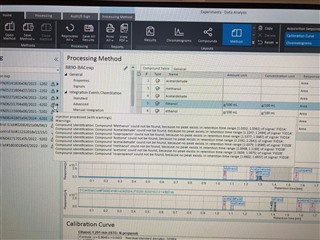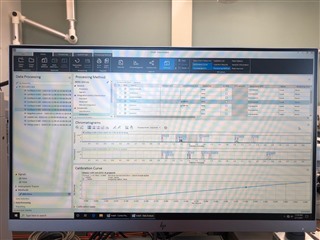
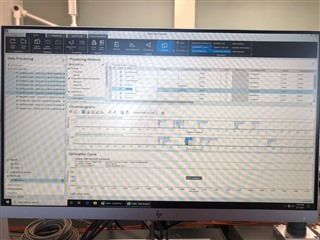
(method partially calibrating on one column and not at all on the other)
Hey all,
I have not been able to figure out why I am getting a partial calibration in real time with my brand new method. I am doing HS-GC-FID dual column analysis.
I have six analyte levels with an internal standard.
I have properly populated the "Calibration" under my processing method with identifying my internal standards and associating my analytes of interest with their proper internal standard. I put in the concentrations of all six levels.
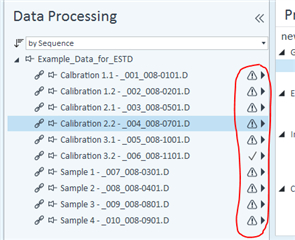
Then, on my sequence I designate my calibration levels of 1-6 and use the "Clear all calibrations" from the dropdown for the first cal.std type so that a new curve should be generated. However, I am only getting a two point calibration (my two highest standards) on only one of my columns. The other column's curve is totally blank.
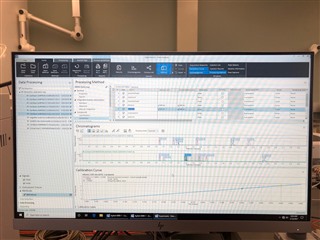
All the peaks/data are present in all six calibrators and when I reprocess that sequence with the method I used in the sequence in the first place, it properly calibrates and all six points are present on both columns! How is this not working in real time, but its working in reprocessing?
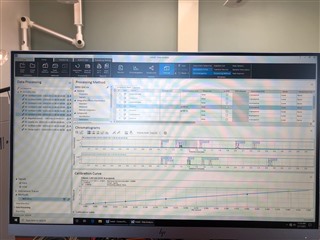
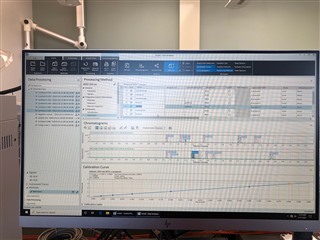
(note successful 6-point calibration from reprocessing of the exact same method)