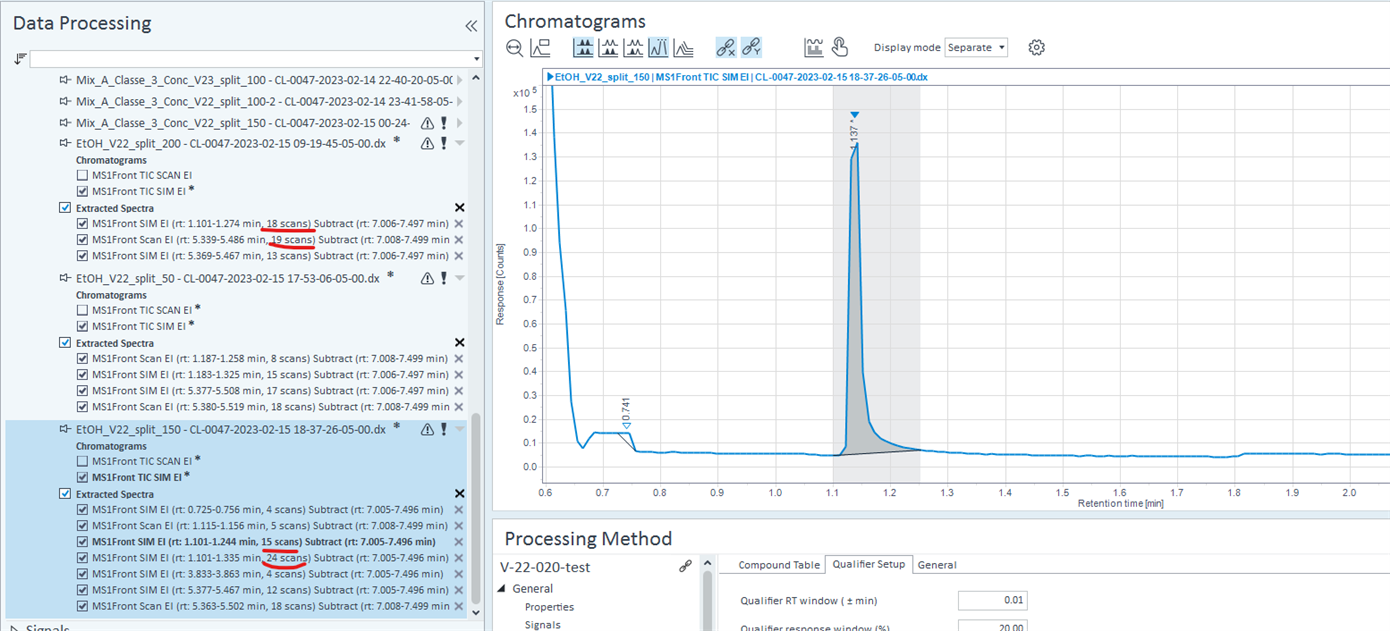
Hi everyone,
I knowhow to extract the number of data (or scan) by peaks with masshunter, but I can I get this information with openlab CDS
thank you
JF Fortin
Hi everyone,
I knowhow to extract the number of data (or scan) by peaks with masshunter, but I can I get this information with openlab CDS
thank you
JF Fortin
Hello,
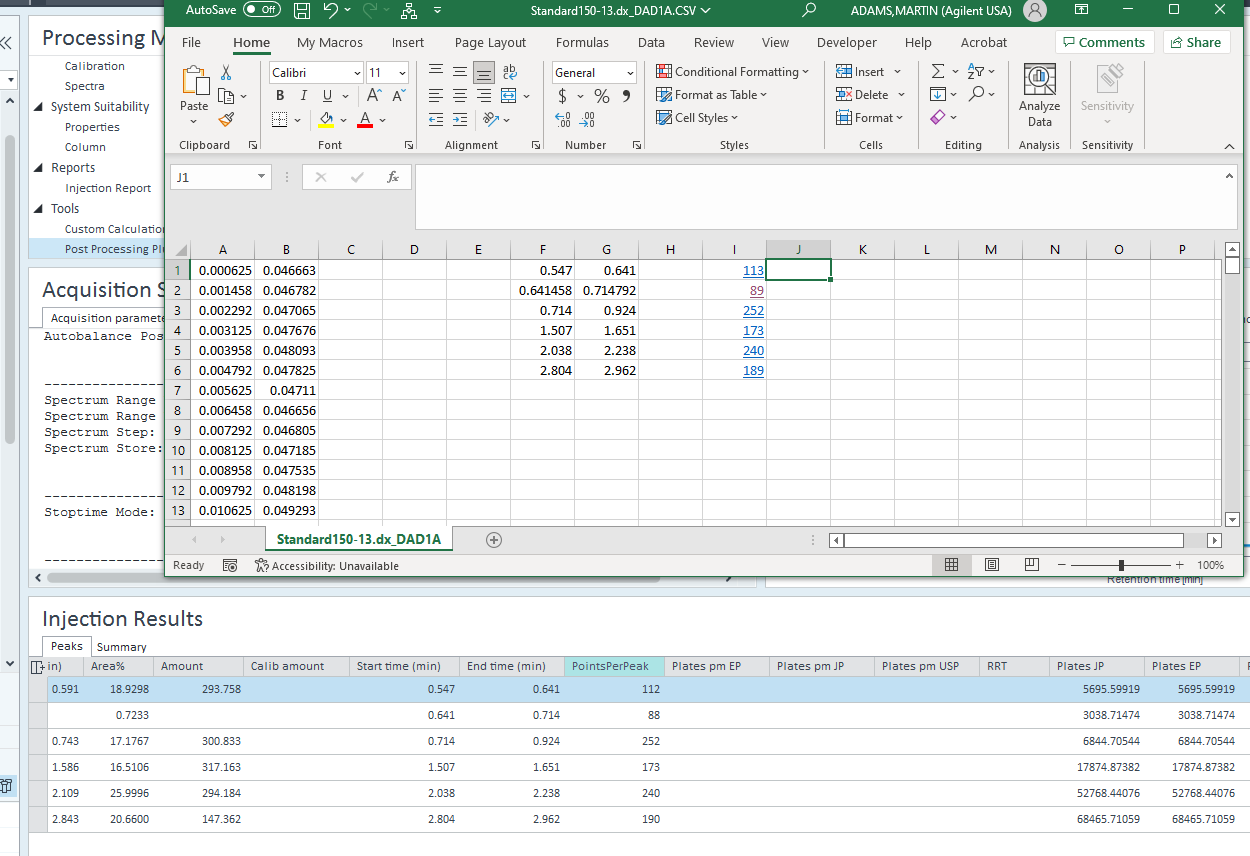
You would need to use a custom calculation in CDS to calculate the data points. As an example, the CC below uses a enter value for data rate in the CCF file to calculate an estimate for the number of points from the peak start and peak end times. You would need to provide the data rate as in the example or using a custom field. This is because the number from the method cannot be accessed for calculations. I compared the calculations to the actual points in excel from the CSV export of the chromatogram.
Marty Adams
(Peak_EndTime - Peak_BeginTime) * 60 * DataRate
Thank you Martin.
You have a lot of points/peak. Usually we want to have around 5-20 points/peaks. Are you sure that the formula is ok?
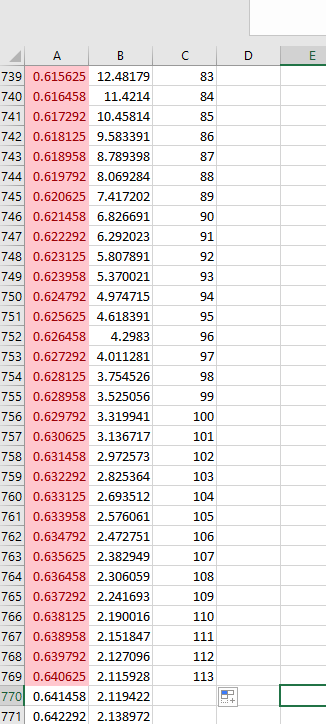
The calculation is just peak end time in minutes - peak start time in minutes, multiplied by 60 to convert to seconds, then multiplied by the data rate in points per second. Also note that I counted the actual points from the x,y data in excel for the same peaks and got approximately the same results. It is approximate since I am using the rounded values for the start and end points in excel. This is just some short column HPLC demo data and is likely over collected at 20Hz.
Marty Adams
Thank you Martin for this precious help. I am looking to get this custom calculation done in near future.Regards
I think I saw where to find the number of scan or point/peak on Openlab CDS similar to what we can use with MassHunter qualitative software. I am asking if it's equivalent with Agilent Canada. I have no answer from them yet.