Hello all,
How do I extract a Spectrum from a Peak in a chromatogram (Openlab CDs software) and How do I compare my sample with a standard?
I would like to know If the sample have the same Spectrum that the standard.
Hello all,
How do I extract a Spectrum from a Peak in a chromatogram (Openlab CDs software) and How do I compare my sample with a standard?
I would like to know If the sample have the same Spectrum that the standard.
Hi julia.jsd thank you for participating in the Agilent Community! I moved your question to the Chromatography Software forum for better visibility.
Would you please let us know what version of OpenLab CDS Software you have?
Thanks,
Kristen
Hello Kristen,
Many thanks to move it. I uses 2.5 version of Openlab CDS.
Hello,
You will need to setup the method to confirm the compound by UV spectrum. See the page from the help below. You can search Confirm compound identity (UV) in the help for more information.
Marty Adams
You can confirm the compound identity by comparing a UV reference spectrum with the current UV spectrum of a peak.
In the injection tree, select a reference injection. This injection contains the compound or compounds that you want to use to create the UV reference spectra.
Add the UV reference spectra to your processing method.
Click Reprocess All.
The spectra at the peak apexes are compared with the UV reference spectra stored in the method. The UV confirmation match factor is calculated.
To show the confirmation match factor: In the Injection Results window, choose the UV Conf. Match Factor column.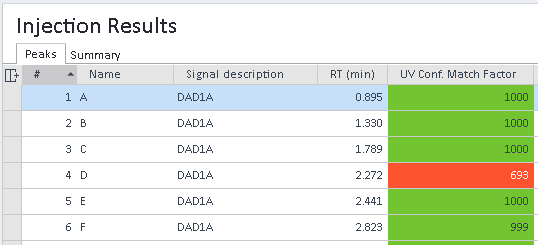
If the match factor is greater than the limit set in the method, the compound identity is confirmed and the match factor is displayed with a green background; otherwise it is displayed with a red background. Generally, match factors can be evaluated as follows:
Match factor > 995: Spectra are very similar.
995 > Match factor > 990: There is some similarity.
Match factor < 990: Data should be observed carefully.