Hi All,
I am aware that this problem has been answered well in this forum but I am not able to solve it for my scenario.
In my analysis, i have two peaks which are the meta- and para-cresol. I want to show the peak area and percentage area of sum of both peaks.
In processing, I grouped both peaks using timed and named. Only timed appeared as below using the group Table template. Not sure why the named did not even show in the processing Tab.
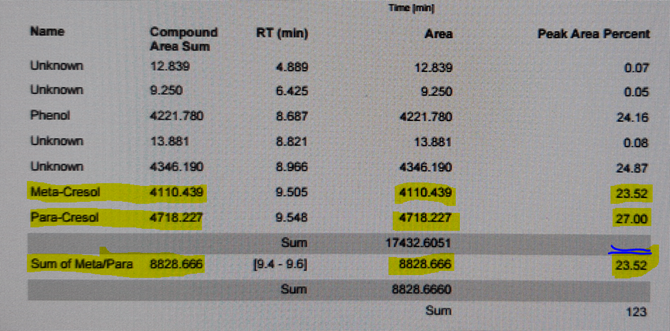
My question is why the peak area % is not the combined value. How to correct it?
Another question is how to not split the merged peak during automatic integration in Openlab CDS 2.3?
I tried by changing the peak width and skin to valley ratio but it is not a good way as it affects the integration of small peaks for other samples.
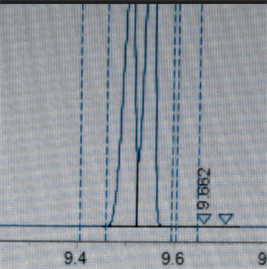
Thank you for your help :)
Regards
Kris
References referred.
How can I add timed group (isomers) in the OpenLAB Intelligent Report?
