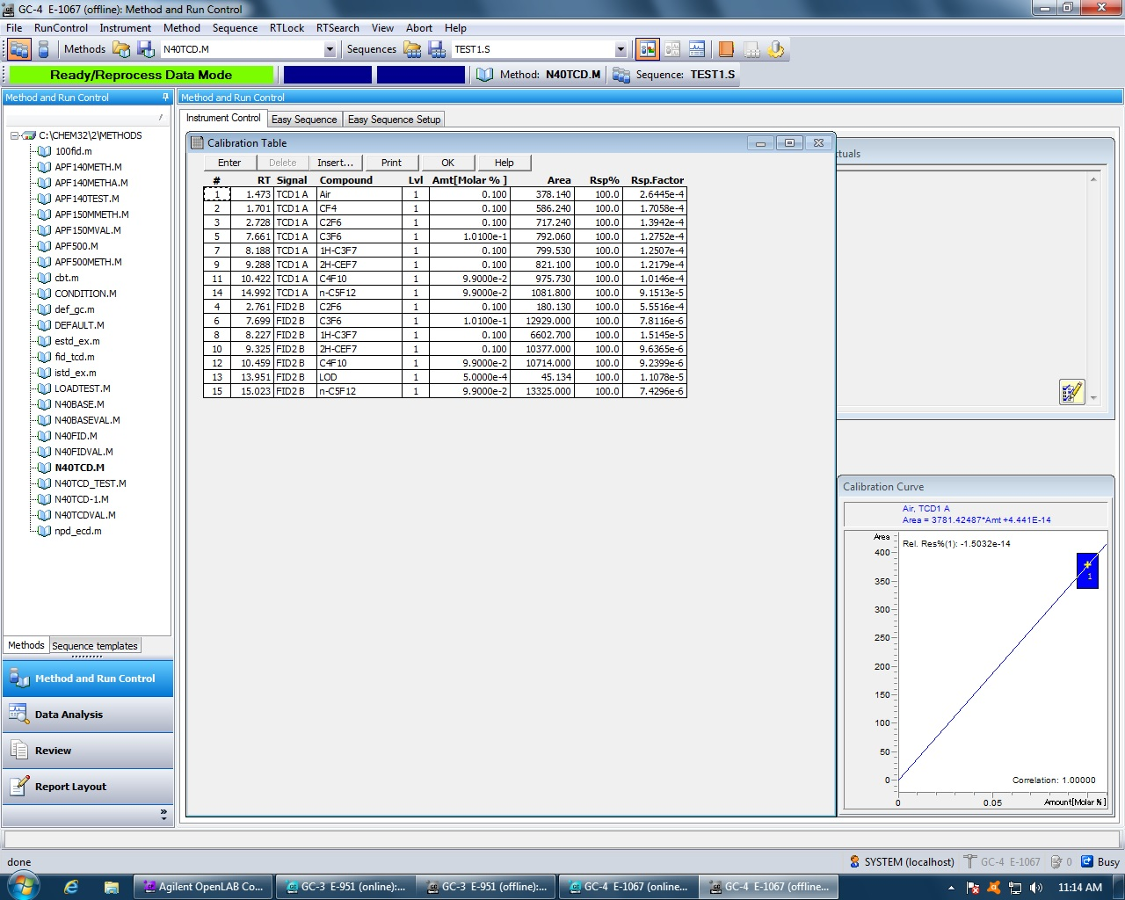
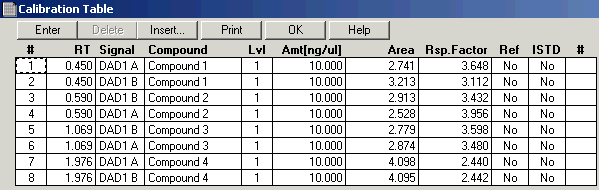
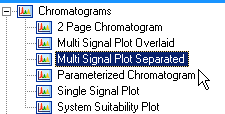
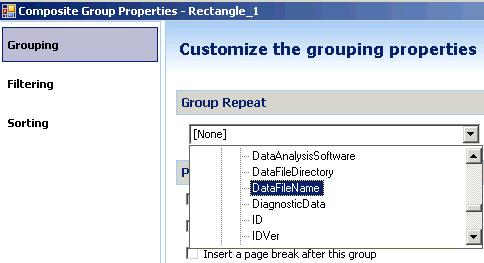
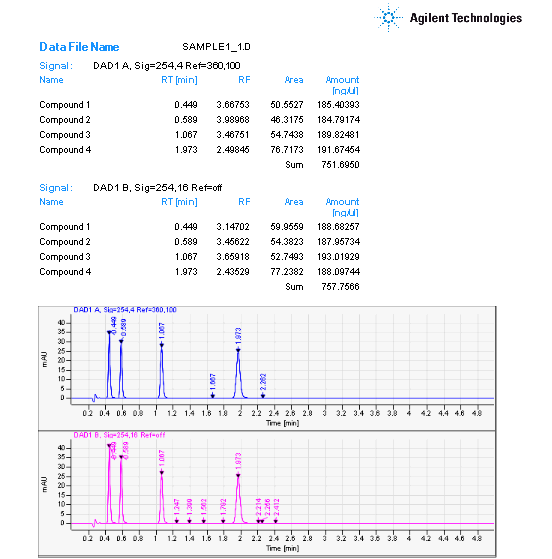
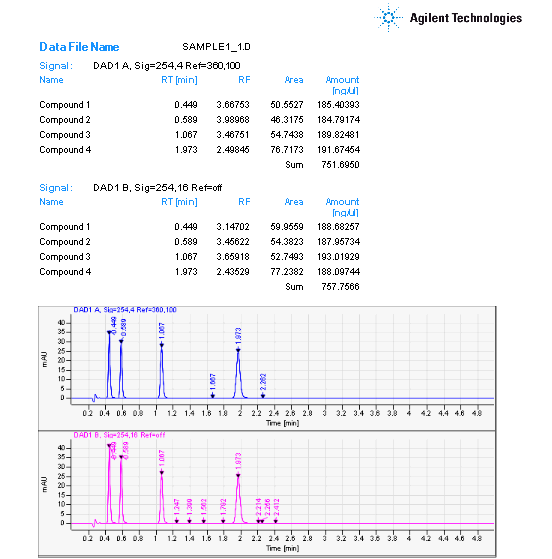
I've tried using the classic report style in OpenLab CDS and creating an intelligent report. Neither one will report a summary report for the FID signal results. I'm running a 7890B with TCD and FID in series. The sample report will print the data for each signal and the data is grouped by detector signal. However, the plots are not separated with the data.
Anyone else have this problem?
Thank you,
Mary Aaron