The MassHunter Productivity App allows a lab manager to add their own methods in addition to the default method. This document will outline some considerations to make when adding custom methods.
1. Method Alignment Error Checking
From the perspective of the Productivity App, a single method consists of an acquisition and analysis method pair. This pairing allows the app to automatically analyze the acquired data. To ensure that all compounds declared in the acquisition method can be properly analyzed by the analysis method, the app will verify the following on import:
- All compounds in the acquisition method are contained in the analysis method
- All transition information is identical between the methods for all compounds in the acquisition method
- All ISTD compounds in the analysis method exist in the acquisition method for all compounds in the acquisition method that have associated ISTDs
These rules allow the analysis method to contain more compounds than the acquisition method because the extraneous compounds will simply be removed upon analysis. If there are any issues with the two methods, a message will display describing the issues and the methods will not be added to the App.
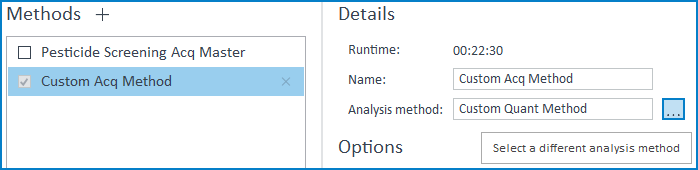
2. Modifying an Added Custom Method
When a method pair is added to the Productivity App, the app creates a private copy of the method files. This ensures that the method referenced by the app cannot be unintentionally modified if the original method is changed or moved. If you do need to modify a method that has already been added to the app, you will need to modify the original method and re-add the modified method to the app. If only the analysis method needs modification, you can simply swap the analysis method from the focused run method (the same validation will apply, see above):
However, if you modify the acquisition method, you will need to remove the old method and re-add both the modified acquisition method and associated analysis method. If all you need to do is adjust the RT values for the method, you can use the RT Tune function offered by the app. For more info, see Tuning Retention Times in the MassHunter Productivity App.
3. ISTD Mapping Import
The analysis method controls ISTD compound mapping. If there is ISTD mapping information, the application will use the mapping for method validation (see above) as well as specialized ISTD compound management. Because ISTD compounds are required purely for analysis and are not a target compound of interest, the app will manage ISTD compounds in the following ways:
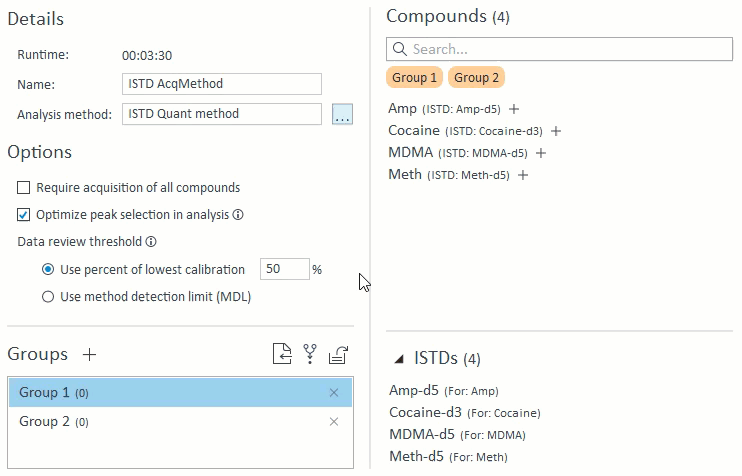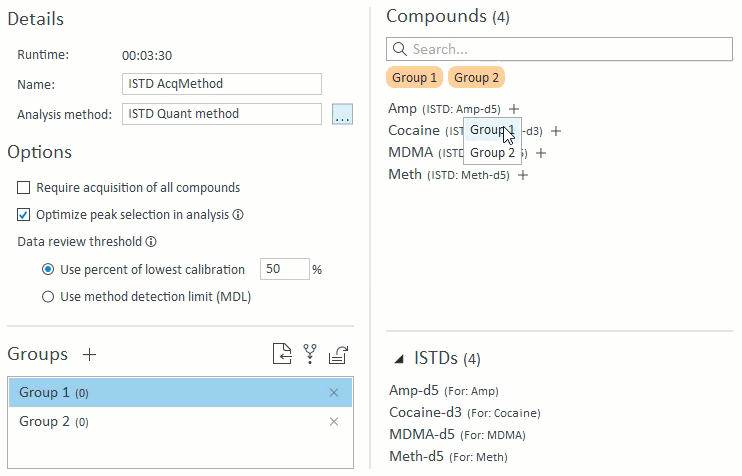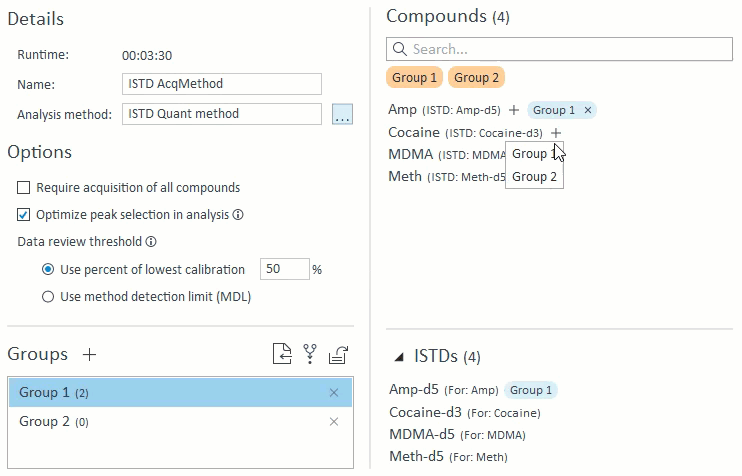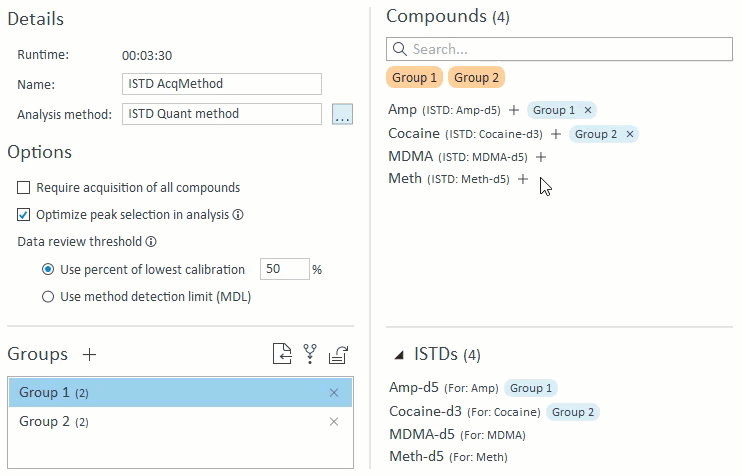
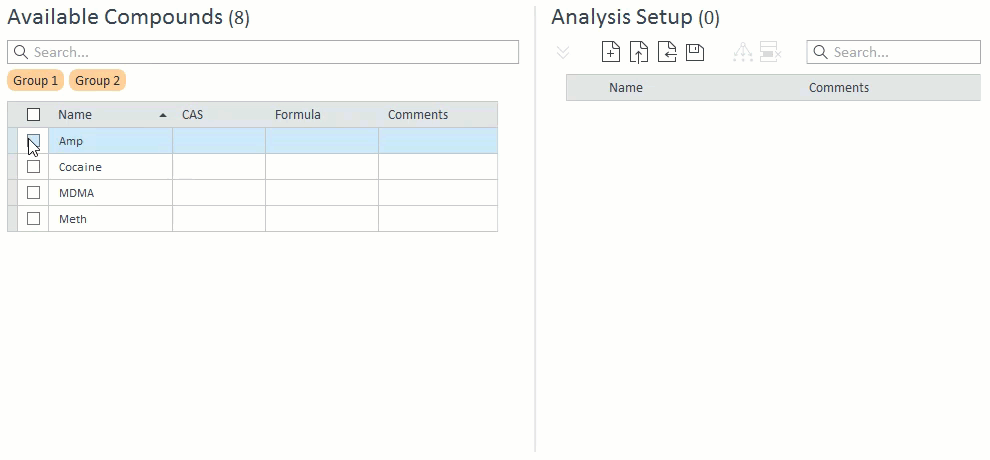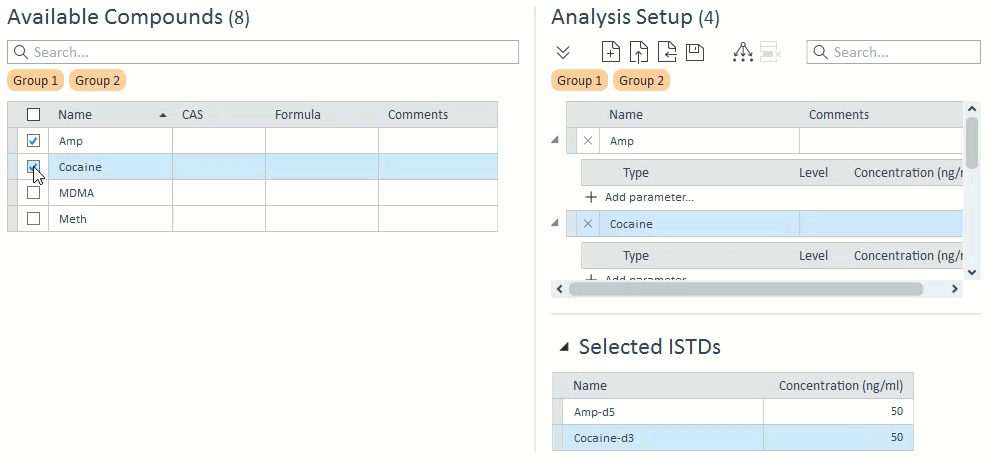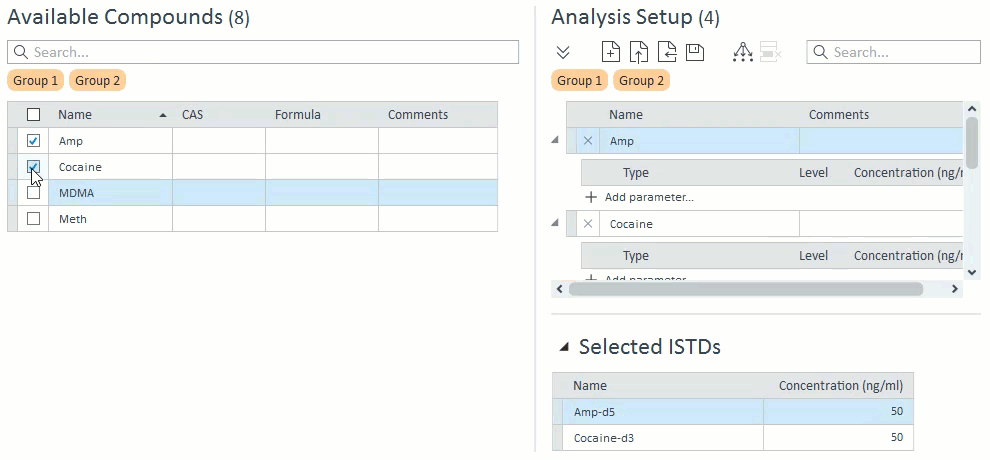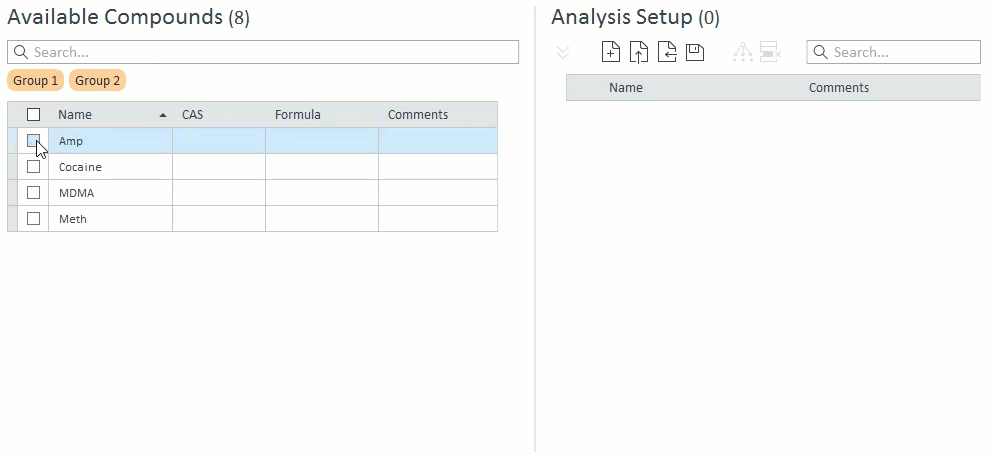
- When managing compound groups, the app will automatically add any ISTD compounds to the group when you add their mapped target compounds:
- When selecting compounds for acquisition and analysis, ISTD compounds will be hidden and cannot be selected on their own. If a target compound is selected that has an ISTD, that ISTD will automatically be selected and will appear in the Analysis Setup panel on the right:
4. Compound Group Import
If compound groups have already been defined in the acquisition method, compound groups will be automatically imported with the method. Names separated by commas will be treated as two separate groups. For more information on how compound groups can be used, see Optimizing Efficiency with Compound Groups in the MassHunter Productivity App.
5. Compound Metadata Import
When a method is added to the App, the compound information is imported so that it can be displayed in the compound selection page. If you want to display CAS numbers and formulas alongside the compound name, you can provide a CSV file titled "CompoundMetadata.csv". This file contains a list of compound names, CAS numbers, and formulas. For an example of this metadata file, see the installed copy of the default acquisition method at <MassHunter Home>\Methods\Productivity App.
Related Links:
Tuning Retention Times in the MassHunter Productivity App
Optimizing Efficiency with Compound Groups in the MassHunter Productivity App