In order to extract a chromatogram at any collected wavelength you could use the procedure below.
Caution: The wavelength specific data density of the UV file is typically smaller compared to a single signal file. This typically results in a lower S/N ratio and it's not recommended to use such extracted signals for quantification.
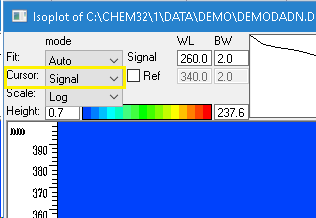
- In Data Analysis view got to menu Spectra > Isoabsorbance plot
- Extract the signal from iso-absorbance by changing the cursor to “signal” and enter the wavelength and bandwidth setting. This will activate the COPY button
- Press the Copy button and close the iso-absorbance plot window
- Integrate the extracted chromatogram
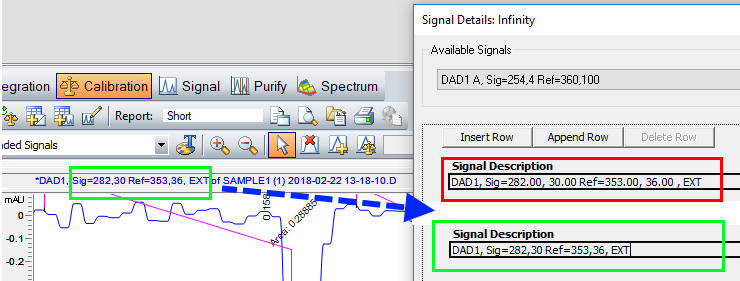
- Calibrate the extracted signal (this action will copy the correct syntax to Calibration > Signal Details)
- Delete the calibration table, in case you don’t need it. The Signal Details are kept.
When loading another data file and the View > Preferences are set to "Load signals using signal details", the chromatogram gets automatically extracted when loading a data file with appropriate spectral data
Note: If you aim to use the calibration table for compound indentification or quantification, you need to ensure the signal description shown in the signal details window matches the signal description in the header of the chromatogram window. For certain UV detectors you need to manually shorten wavelength numbers as shown below. After all, quantifications based on extracted signals need to evaluated very carefully due to the reduced signal/noise ratio and its impact on quantification precision.