Hello,
I have two questions about the report section in OpenLab CDS:
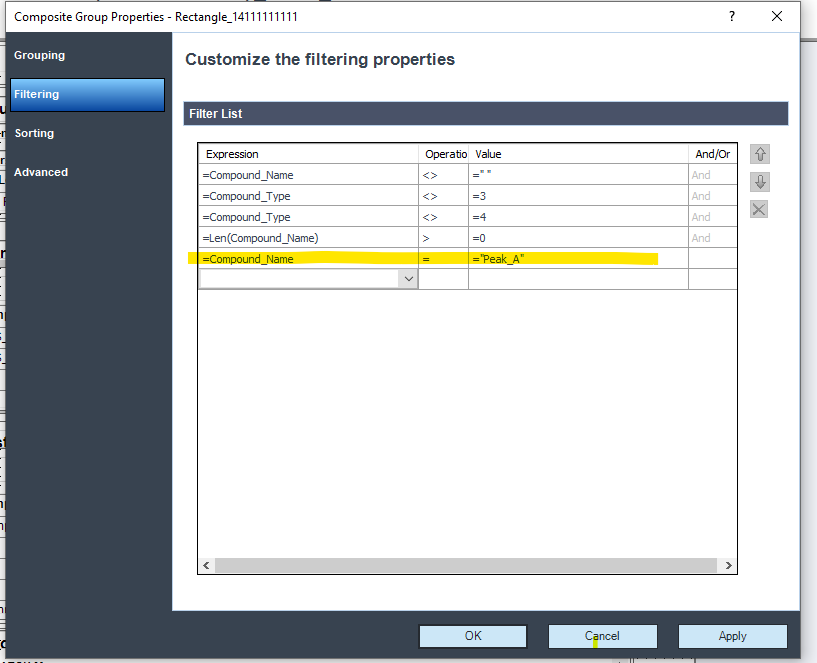
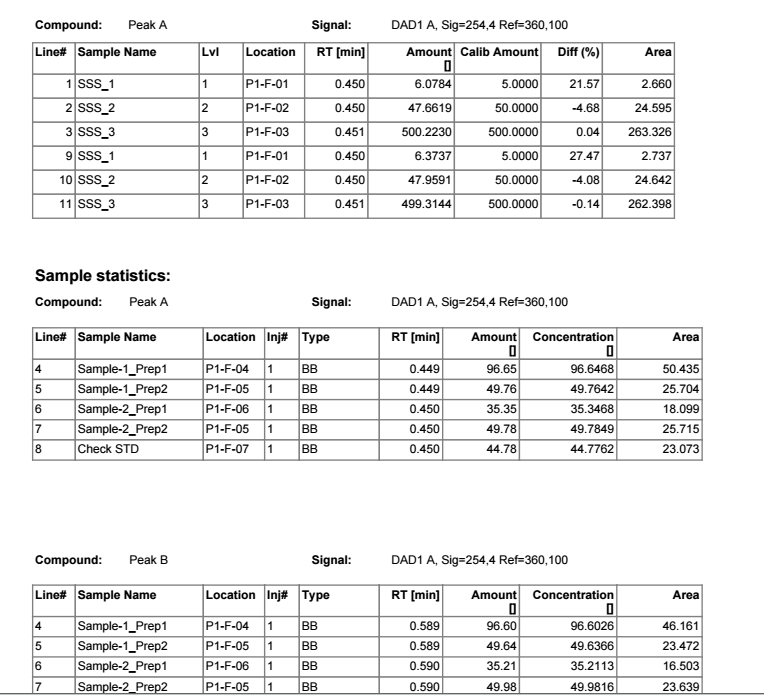
1- Is there a way to set the report for one compound out of multiple compounds?
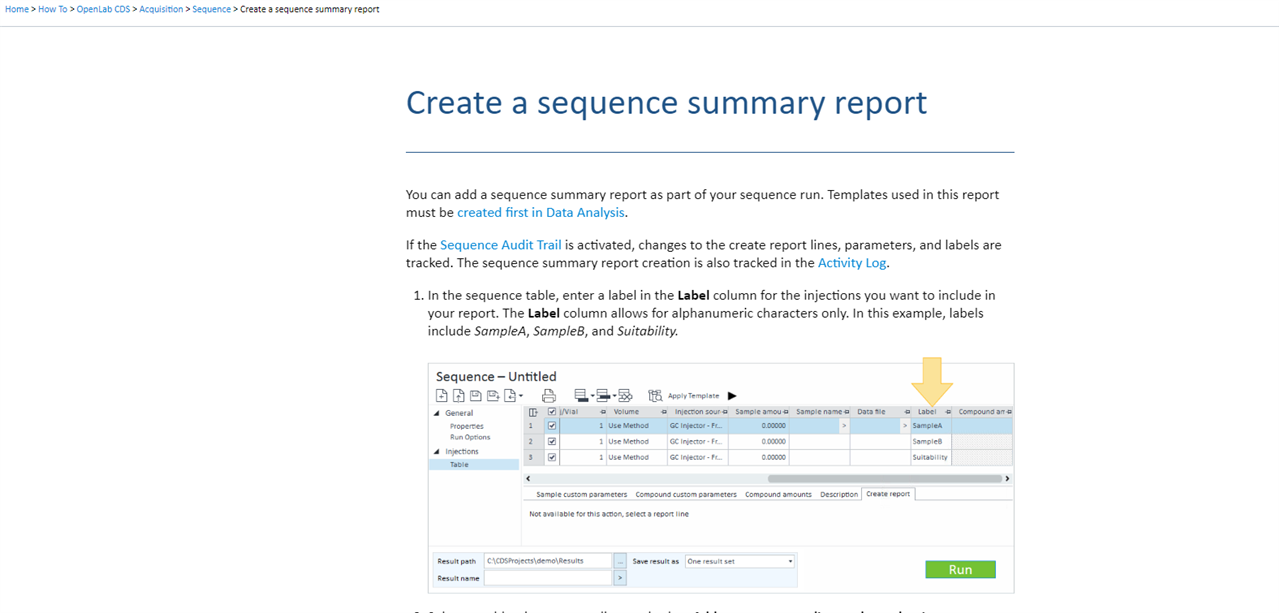
2- is there a way to get the sequence result at the end as one PDF file rather than having multiple injections results separately as individual PDF files?
Thank you,
-Sam