Hello,
I am very new to OpenLab CDS. I am trying to to create a calibration curve using an internal standard (1-pentadecanol) to find the assay of Cetyl Alcohol using a USP Cetyl Alcohol Reference Standard as my calibration standard. I have been trying to get help using the 'Getting Started' section but I am still struggling a little about how this is done.
Concentration of Cetyl Alcohol in Std = 53.74mg/50mL (1.0748mg/mL)
Concentration of 1-pentadecanol Int Std = 500.28mg/500mL (1.00056mg/mL)
Unknown Sample weight = 51.96mg/50mL
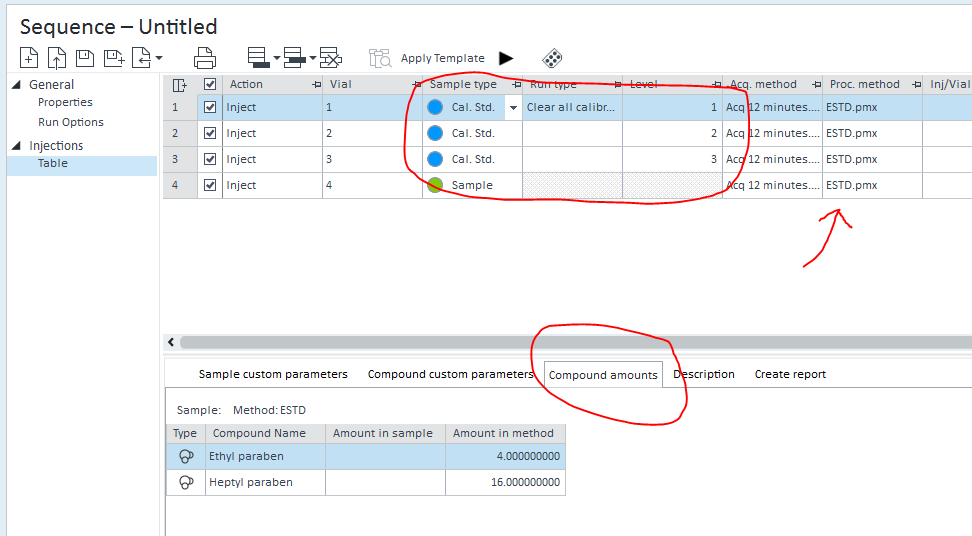
The run sequence consists of 5 standard injections followed by the sample injection which is then followed by a single bracketing standard injection. I used the "Overall" bracketing mode to bracket the sample with the calibrating standard injections. Once the Acquisition was over and I started Data Analysis, I loaded the injections and linked them to the Processing Method. In my Processing Method Compound Table under the Calibration Node, I have been able to mark 1-pentadecanol as the ISTD by checking the "Is ISTD" box. I have also selected "Associated ISTD" as 1-pentadecanol for the Cetyl Alcohol line in the compound table. In the 'General' tab I selected "Internal Standard", Number of Levels: 1 and Curve Calculation: From Average per Level. The RF definition is Response per amount. I also selected "Calculate Mass%". The question I have is -
1. Do I enter the ISTD amount in the processing method? or do I enter it in the injection list? Both?
2. I am assuming that by 'Amount' the software means concentration of the 1-pentadecanol (i.e. 1.00056mg/mL) or am I wrong and it actually wants me to enter the weight (i.e. 500.28mg)
3. Under the Compound Amounts column in the Injection List, I entered the concentration of my calibration standard (i.e. 1.0748mg/mL) for each line of the calibrated standard. Is this correct? I had to do this after the acquisition was finished because at the time of the acquisition, I still did not have a processing method to enter in the sequence, so now there is a 'modified' under the compound amounts column in the injection list which I have to click to open a smaller window on the right where I have to enter the amount.
4. In the Injection List, on the sample line I entered the weight of my sample (i.e. 519.6mg) under the column for Sample Amount. All the Internal Standard Amount lines in the Injection List were 1.000 because I had entered the Internal Standard Amount in the processing method compound table.
After doing all this I selected all injections and linked them to my processing method and reprocessed. I changed to the "Results" tab to see the results and the result under the 'Amount' column of the Sample line in table is 1.012 which is not even close to what I was expecting. I was expecting a number close too 100 (like 98.26 or something like that) for the assay result. If I do the calculations by hand using the response numbers from the injections I get the right answer so how can I get the software to work for me.
Can anyone please provide some insight and answers. I will appreciate it very much.