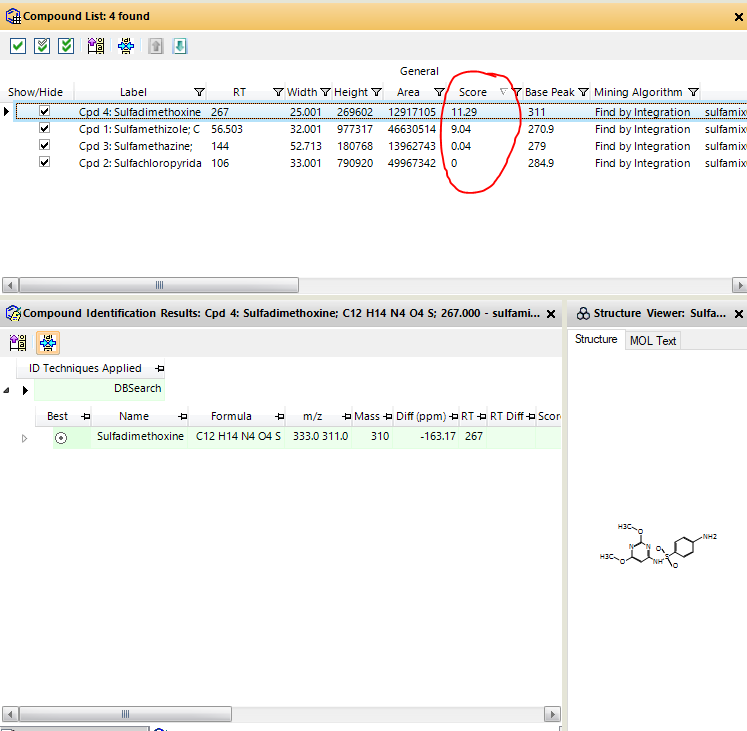
Hello!
I'm new to the MassHunter Qualitative Analysis software and I'm hoping someone can help me to understand some information. After matching the compounds with the existing library/database, the compound information is presented under the "Compounds List" tab. What is the meaning of the numbers in the "Score" column? Is there a way to find the sample purity or composition?
Thank you for your help.