Dear all,
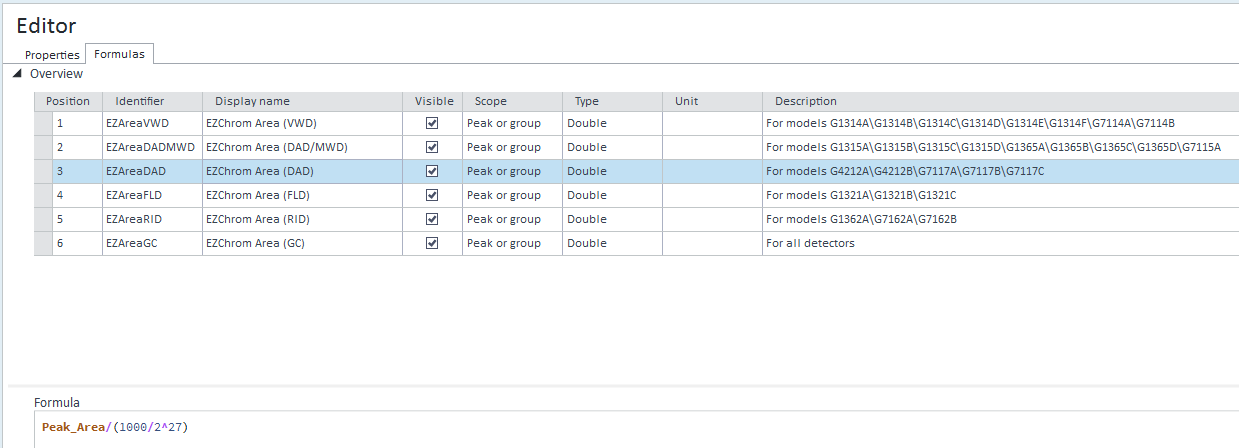
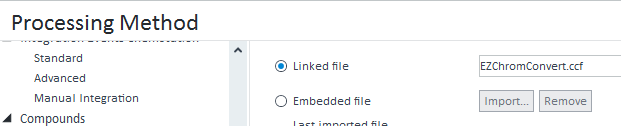
Just quick help needed, basically we are using Ezchrom wherein the area response is in millions but last year we moved to open lab CDS2.3 version, in this software with the same concentration of sample and the area response comes in thousands which have big impact in term of uncertainty between Ezchrom and open-lab because in our other lab they are still using Ezchrom but not Open-lab.I am worry about method transfer. Can you please guide me how to set the area response in open lab CDS 2.3 version.