(Running OpenLAB CDS Chemstation Edition, version C.01.07 SR1; with ECM)
We have written into our SOPs that the data file and the sample name must match. This was done to provide another layer of data integrity. We also have a rule that system suitability must be verified as passing prior to injecting any samples. This leads us to create a lot of sequences that have two parts: system suitability and samples, that have to be combined into a single result set.
All of this causes us two problems:
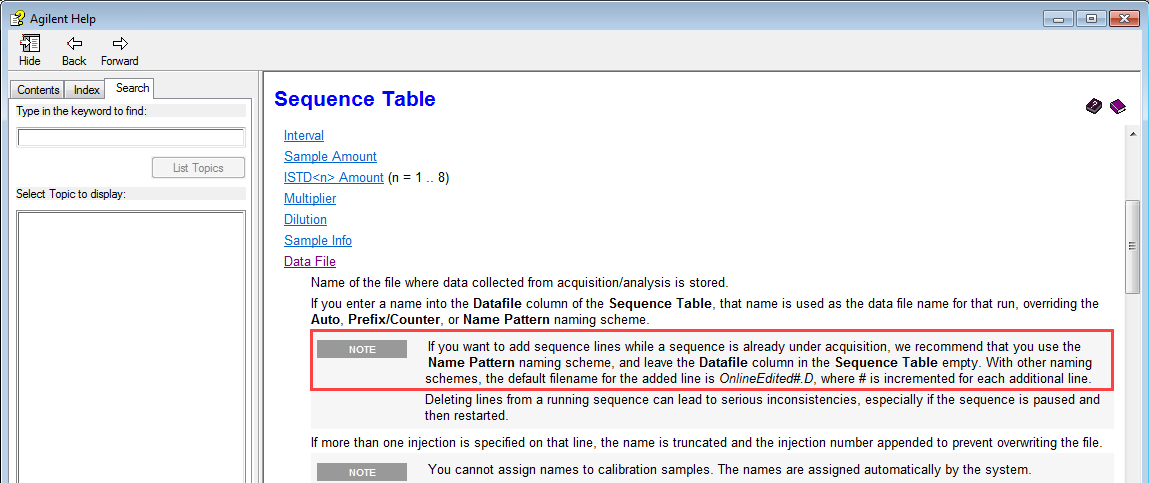
1) The dreaded "OnlineEdited" tag on datafiles that are, well, edited online, while the sequence is running. This occurs when an analyst uses the offline to verify that their running sequence does meet system suitability and decides to add their samples to the online sequence. But not every time does it get tagged as Online Edited. Does anyone know what, exactly, triggers the OnlineEdited tag?
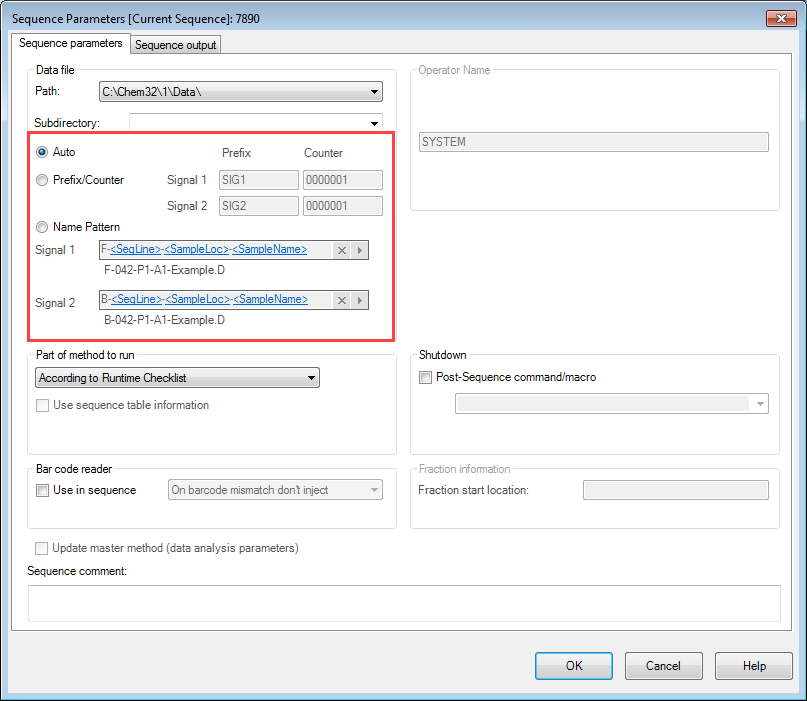
2) If the Sequence Parameters aren't set properly, the Sample Name doesn't get added to the data file name. We've recently add multiple instances of sequence tables where the default selection in the parameters seems to have changed to "Auto" instead of "Name Pattern". The analysts say they didn't change anything (I tend to believe them), but every time I create a sequence, the Name Pattern option is automatically selected. Does anyone know how to change / verify the selection for the default sequence template?
We also have a problem where the analyst wants to define everything about the sample in the sample name field (e.g. product, lot, manufacturing stage, replicate prep number, stability conditions, etc.) and most of it gets cut off. I've told them this is because Windows limits us to 256 characters in the file location and they're using too many of them. That information should go in the sample description instead. (But no one seems to want to hear that.)
Thanks in advance for your help.