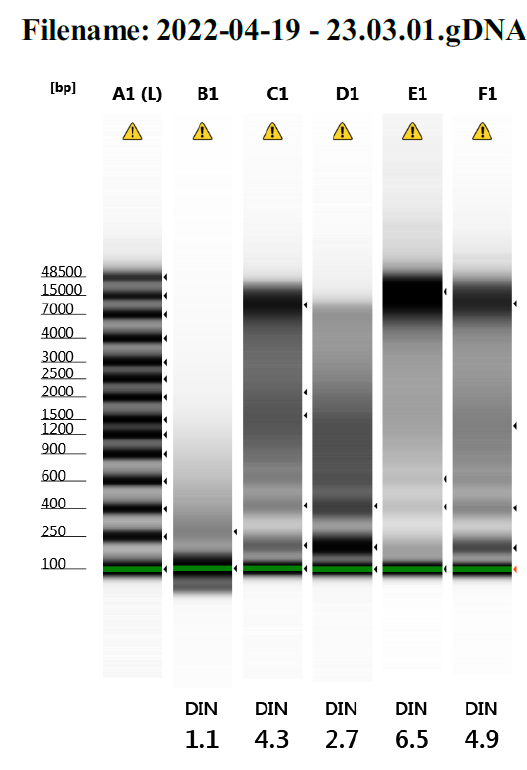
We have tried several times to isolate genomic DNA from fish fin clips using proteinase K and have run the samples on the TapeStation. Sometimes we get high molecular weight DNA, but other times we get what looks like degraded DNA with a series of low molecular weight bands. It is hard for me to imagine that these consistent bands are produced by DNA degradation. Does anyone have any thought on what these are? Thank you for your help.